
Apis mellifera associated microvirus 38
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.95
Get precalculated fractions of proteins
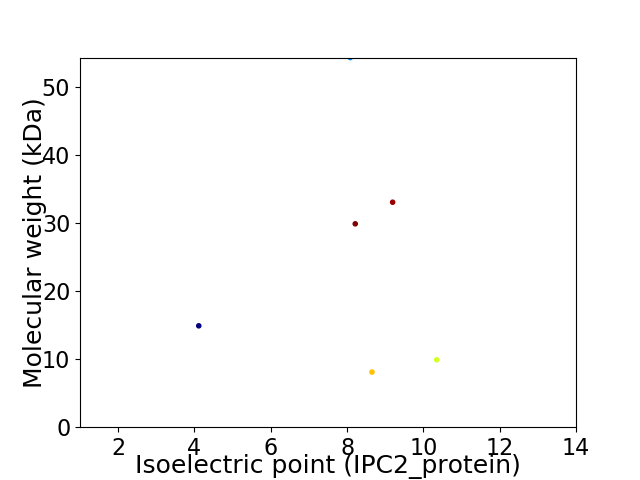
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8UTI8|A0A3S8UTI8_9VIRU Uncharacterized protein OS=Apis mellifera associated microvirus 38 OX=2494767 PE=4 SV=1
MM1 pKa = 7.54KK2 pKa = 10.37VFQNIEE8 pKa = 3.8TGEE11 pKa = 4.32IITTHH16 pKa = 6.82DD17 pKa = 3.47KK18 pKa = 10.84FGQEE22 pKa = 4.46LPDD25 pKa = 4.33PTPMAPPVGYY35 pKa = 10.23VRR37 pKa = 11.84QPSLAEE43 pKa = 3.8QIRR46 pKa = 11.84NMVRR50 pKa = 11.84SQHH53 pKa = 5.55LAMVAEE59 pKa = 4.37SQGFEE64 pKa = 4.21TFEE67 pKa = 4.03EE68 pKa = 4.81ADD70 pKa = 4.47DD71 pKa = 5.34LDD73 pKa = 5.02IPDD76 pKa = 5.31DD77 pKa = 4.32VDD79 pKa = 4.48PSSPWEE85 pKa = 4.05EE86 pKa = 4.48VYY88 pKa = 10.64EE89 pKa = 4.3PPLPPGNITPLEE101 pKa = 4.01EE102 pKa = 5.6AISQAKK108 pKa = 9.14ARR110 pKa = 11.84QPAQSPPEE118 pKa = 4.09PLSAGGASPPAPTSPPPEE136 pKa = 5.22GGAA139 pKa = 3.53
MM1 pKa = 7.54KK2 pKa = 10.37VFQNIEE8 pKa = 3.8TGEE11 pKa = 4.32IITTHH16 pKa = 6.82DD17 pKa = 3.47KK18 pKa = 10.84FGQEE22 pKa = 4.46LPDD25 pKa = 4.33PTPMAPPVGYY35 pKa = 10.23VRR37 pKa = 11.84QPSLAEE43 pKa = 3.8QIRR46 pKa = 11.84NMVRR50 pKa = 11.84SQHH53 pKa = 5.55LAMVAEE59 pKa = 4.37SQGFEE64 pKa = 4.21TFEE67 pKa = 4.03EE68 pKa = 4.81ADD70 pKa = 4.47DD71 pKa = 5.34LDD73 pKa = 5.02IPDD76 pKa = 5.31DD77 pKa = 4.32VDD79 pKa = 4.48PSSPWEE85 pKa = 4.05EE86 pKa = 4.48VYY88 pKa = 10.64EE89 pKa = 4.3PPLPPGNITPLEE101 pKa = 4.01EE102 pKa = 5.6AISQAKK108 pKa = 9.14ARR110 pKa = 11.84QPAQSPPEE118 pKa = 4.09PLSAGGASPPAPTSPPPEE136 pKa = 5.22GGAA139 pKa = 3.53
Molecular weight: 14.91 kDa
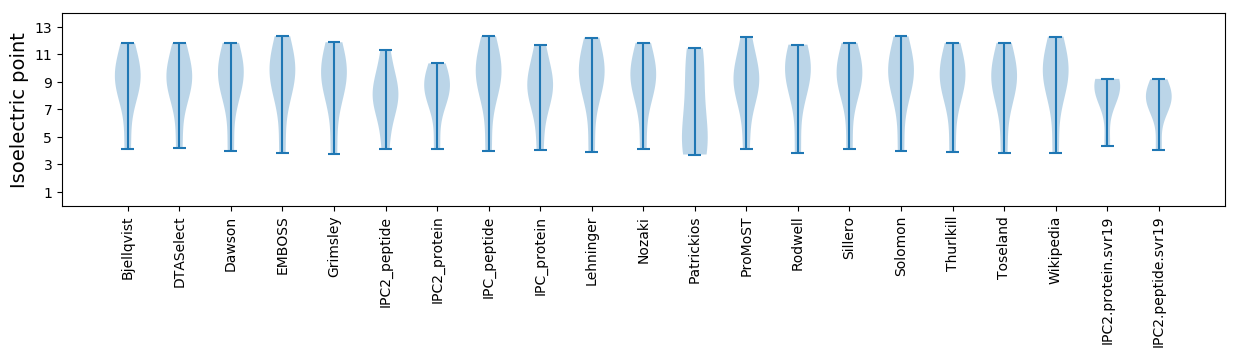
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UTL9|A0A3S8UTL9_9VIRU DNA pilot protein OS=Apis mellifera associated microvirus 38 OX=2494767 PE=4 SV=1
MM1 pKa = 7.7RR2 pKa = 11.84CANPYY7 pKa = 8.39TGSGGMFGCGQCLPCRR23 pKa = 11.84FNRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84TWQHH32 pKa = 5.33RR33 pKa = 11.84LILEE37 pKa = 4.62ATQHH41 pKa = 4.38SHH43 pKa = 4.27QTFVTLTYY51 pKa = 10.76DD52 pKa = 3.74DD53 pKa = 3.76EE54 pKa = 4.75HH55 pKa = 8.37VPEE58 pKa = 4.38GHH60 pKa = 6.01TLVPKK65 pKa = 10.42HH66 pKa = 5.99LQDD69 pKa = 3.25FLKK72 pKa = 10.52RR73 pKa = 11.84LRR75 pKa = 11.84HH76 pKa = 5.12KK77 pKa = 10.16QQIRR81 pKa = 11.84YY82 pKa = 8.14FAVGEE87 pKa = 4.16YY88 pKa = 11.11GEE90 pKa = 4.57TTQRR94 pKa = 11.84PHH96 pKa = 5.1YY97 pKa = 9.83HH98 pKa = 6.58LALFGYY104 pKa = 7.63PNCNWLQSRR113 pKa = 11.84YY114 pKa = 7.48AHH116 pKa = 7.49RR117 pKa = 11.84KK118 pKa = 7.26RR119 pKa = 11.84CCVACDD125 pKa = 4.23TIRR128 pKa = 11.84DD129 pKa = 3.33TWGRR133 pKa = 11.84GNVFLGTLSSDD144 pKa = 3.45SASYY148 pKa = 9.58VAGYY152 pKa = 9.19VCKK155 pKa = 10.71KK156 pKa = 6.51MTNKK160 pKa = 10.15DD161 pKa = 3.6DD162 pKa = 3.55PRR164 pKa = 11.84LAGRR168 pKa = 11.84HH169 pKa = 5.46PEE171 pKa = 4.19FSRR174 pKa = 11.84MSLKK178 pKa = 10.62PGIGFHH184 pKa = 6.7ALAKK188 pKa = 10.08IADD191 pKa = 4.89AIRR194 pKa = 11.84LNKK197 pKa = 9.51IDD199 pKa = 4.15TLLGDD204 pKa = 3.79VPASLRR210 pKa = 11.84HH211 pKa = 5.92GSRR214 pKa = 11.84IMPLGRR220 pKa = 11.84YY221 pKa = 8.45LRR223 pKa = 11.84SQLRR227 pKa = 11.84LAVMGQKK234 pKa = 9.75NASPEE239 pKa = 3.96AQIKK243 pKa = 10.34AGEE246 pKa = 4.13ALHH249 pKa = 5.81TLRR252 pKa = 11.84ILAKK256 pKa = 9.33ALNYY260 pKa = 9.67EE261 pKa = 4.34KK262 pKa = 10.34EE263 pKa = 4.07VHH265 pKa = 5.95EE266 pKa = 5.01LIKK269 pKa = 11.0EE270 pKa = 4.3EE271 pKa = 4.47IGNTAANAAMKK282 pKa = 10.43HH283 pKa = 5.56KK284 pKa = 9.33LTNRR288 pKa = 11.84RR289 pKa = 11.84SLL291 pKa = 3.44
MM1 pKa = 7.7RR2 pKa = 11.84CANPYY7 pKa = 8.39TGSGGMFGCGQCLPCRR23 pKa = 11.84FNRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84TWQHH32 pKa = 5.33RR33 pKa = 11.84LILEE37 pKa = 4.62ATQHH41 pKa = 4.38SHH43 pKa = 4.27QTFVTLTYY51 pKa = 10.76DD52 pKa = 3.74DD53 pKa = 3.76EE54 pKa = 4.75HH55 pKa = 8.37VPEE58 pKa = 4.38GHH60 pKa = 6.01TLVPKK65 pKa = 10.42HH66 pKa = 5.99LQDD69 pKa = 3.25FLKK72 pKa = 10.52RR73 pKa = 11.84LRR75 pKa = 11.84HH76 pKa = 5.12KK77 pKa = 10.16QQIRR81 pKa = 11.84YY82 pKa = 8.14FAVGEE87 pKa = 4.16YY88 pKa = 11.11GEE90 pKa = 4.57TTQRR94 pKa = 11.84PHH96 pKa = 5.1YY97 pKa = 9.83HH98 pKa = 6.58LALFGYY104 pKa = 7.63PNCNWLQSRR113 pKa = 11.84YY114 pKa = 7.48AHH116 pKa = 7.49RR117 pKa = 11.84KK118 pKa = 7.26RR119 pKa = 11.84CCVACDD125 pKa = 4.23TIRR128 pKa = 11.84DD129 pKa = 3.33TWGRR133 pKa = 11.84GNVFLGTLSSDD144 pKa = 3.45SASYY148 pKa = 9.58VAGYY152 pKa = 9.19VCKK155 pKa = 10.71KK156 pKa = 6.51MTNKK160 pKa = 10.15DD161 pKa = 3.6DD162 pKa = 3.55PRR164 pKa = 11.84LAGRR168 pKa = 11.84HH169 pKa = 5.46PEE171 pKa = 4.19FSRR174 pKa = 11.84MSLKK178 pKa = 10.62PGIGFHH184 pKa = 6.7ALAKK188 pKa = 10.08IADD191 pKa = 4.89AIRR194 pKa = 11.84LNKK197 pKa = 9.51IDD199 pKa = 4.15TLLGDD204 pKa = 3.79VPASLRR210 pKa = 11.84HH211 pKa = 5.92GSRR214 pKa = 11.84IMPLGRR220 pKa = 11.84YY221 pKa = 8.45LRR223 pKa = 11.84SQLRR227 pKa = 11.84LAVMGQKK234 pKa = 9.75NASPEE239 pKa = 3.96AQIKK243 pKa = 10.34AGEE246 pKa = 4.13ALHH249 pKa = 5.81TLRR252 pKa = 11.84ILAKK256 pKa = 9.33ALNYY260 pKa = 9.67EE261 pKa = 4.34KK262 pKa = 10.34EE263 pKa = 4.07VHH265 pKa = 5.95EE266 pKa = 5.01LIKK269 pKa = 11.0EE270 pKa = 4.3EE271 pKa = 4.47IGNTAANAAMKK282 pKa = 10.43HH283 pKa = 5.56KK284 pKa = 9.33LTNRR288 pKa = 11.84RR289 pKa = 11.84SLL291 pKa = 3.44
Molecular weight: 33.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1360 |
76 |
493 |
226.7 |
25.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.412 ± 0.23 | 1.103 ± 0.57 |
4.338 ± 0.324 | 5.074 ± 0.672 |
3.456 ± 0.395 | 8.382 ± 0.584 |
3.162 ± 0.818 | 4.559 ± 0.264 |
4.265 ± 0.756 | 7.868 ± 0.731 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.353 ± 0.101 | 4.191 ± 0.342 |
7.5 ± 1.58 | 4.559 ± 1.155 |
7.426 ± 0.871 | 6.691 ± 0.681 |
5.956 ± 0.458 | 5.147 ± 0.722 |
1.324 ± 0.212 | 3.235 ± 0.487 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
