
Changjiang tombus-like virus 6
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.61
Get precalculated fractions of proteins
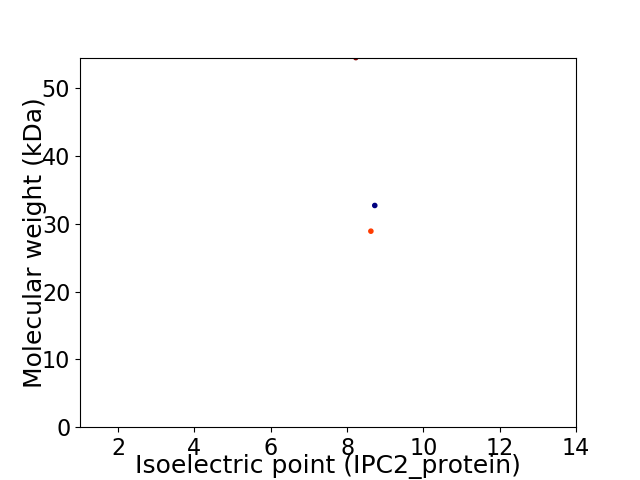
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KG36|A0A1L3KG36_9VIRU RNA-directed RNA polymerase OS=Changjiang tombus-like virus 6 OX=1922820 PE=4 SV=1
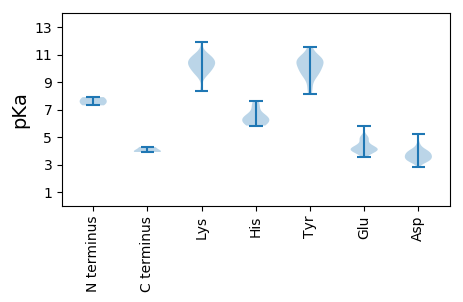
MM1 pKa = 7.3HH2 pKa = 7.64PVFPGLSGALRR13 pKa = 11.84FGVHH17 pKa = 6.25NNNLRR22 pKa = 11.84NGCRR26 pKa = 11.84ALVEE30 pKa = 4.2RR31 pKa = 11.84VFLRR35 pKa = 11.84EE36 pKa = 4.14DD37 pKa = 3.43NGNLIPPPPCVEE49 pKa = 4.46DD50 pKa = 3.46VNKK53 pKa = 9.55TLEE56 pKa = 4.03EE57 pKa = 4.01FKK59 pKa = 10.86AAVITRR65 pKa = 11.84VGVHH69 pKa = 5.88RR70 pKa = 11.84RR71 pKa = 11.84WSVDD75 pKa = 2.8KK76 pKa = 10.69FVNSYY81 pKa = 8.12TGRR84 pKa = 11.84RR85 pKa = 11.84QNVYY89 pKa = 10.58ARR91 pKa = 11.84AAKK94 pKa = 10.23SLEE97 pKa = 4.06TTPVHH102 pKa = 6.0TGDD105 pKa = 3.97FKK107 pKa = 11.88VMNAFVKK114 pKa = 10.65AEE116 pKa = 4.07KK117 pKa = 10.13TNLTSKK123 pKa = 10.16PDD125 pKa = 3.68PAPRR129 pKa = 11.84VIQPRR134 pKa = 11.84NPRR137 pKa = 11.84YY138 pKa = 8.64NVEE141 pKa = 3.56VGRR144 pKa = 11.84FLKK147 pKa = 10.79GLEE150 pKa = 4.05HH151 pKa = 6.66KK152 pKa = 10.39VYY154 pKa = 10.61KK155 pKa = 10.4AVEE158 pKa = 4.73DD159 pKa = 3.63IFGDD163 pKa = 3.61PTVMKK168 pKa = 9.99GYY170 pKa = 9.64NAEE173 pKa = 3.99EE174 pKa = 4.42VAEE177 pKa = 4.17HH178 pKa = 6.42MVSKK182 pKa = 9.3WQSFSDD188 pKa = 4.06PVAVGLDD195 pKa = 3.17ASRR198 pKa = 11.84FDD200 pKa = 3.35QHH202 pKa = 6.42VRR204 pKa = 11.84PSMLEE209 pKa = 3.93WEE211 pKa = 4.36HH212 pKa = 7.3SVYY215 pKa = 10.78AGCFSTGEE223 pKa = 3.93AKK225 pKa = 10.4LLKK228 pKa = 9.68WLLQGQIRR236 pKa = 11.84NVCTMRR242 pKa = 11.84VTDD245 pKa = 4.03GLVKK249 pKa = 10.68YY250 pKa = 9.92RR251 pKa = 11.84VDD253 pKa = 3.56GSRR256 pKa = 11.84MSGDD260 pKa = 3.28MNTALGNCLIMCALVWRR277 pKa = 11.84LGKK280 pKa = 10.52VLGIRR285 pKa = 11.84LKK287 pKa = 10.72LANNGDD293 pKa = 3.83DD294 pKa = 3.31CVVFMEE300 pKa = 4.96RR301 pKa = 11.84KK302 pKa = 10.13DD303 pKa = 3.37LTAFNNKK310 pKa = 8.35VKK312 pKa = 10.61GFFLDD317 pKa = 3.29YY318 pKa = 10.67GFTMKK323 pKa = 10.45VEE325 pKa = 4.17APSFFIEE332 pKa = 4.95QIEE335 pKa = 4.55FCQAKK340 pKa = 9.42PVWDD344 pKa = 3.75GCKK347 pKa = 9.59WIMVRR352 pKa = 11.84DD353 pKa = 3.86PRR355 pKa = 11.84VCTSKK360 pKa = 11.04DD361 pKa = 3.14ATCVVKK367 pKa = 10.62DD368 pKa = 3.58YY369 pKa = 10.91GYY371 pKa = 11.48GSAASYY377 pKa = 9.86WLGAVGEE384 pKa = 4.99CGLAMAGGIPVMQNYY399 pKa = 9.36YY400 pKa = 10.61SAFARR405 pKa = 11.84FGNTNRR411 pKa = 11.84QVQCVTEE418 pKa = 3.85TGMAYY423 pKa = 10.65LARR426 pKa = 11.84GLHH429 pKa = 6.21RR430 pKa = 11.84EE431 pKa = 4.07ATVISADD438 pKa = 3.12ARR440 pKa = 11.84VSFWRR445 pKa = 11.84AFGIAPTQQRR455 pKa = 11.84EE456 pKa = 4.17LEE458 pKa = 4.23KK459 pKa = 10.48WCDD462 pKa = 3.57NTSLTLPNSPCIRR475 pKa = 11.84AYY477 pKa = 9.29PPRR480 pKa = 11.84GILPLLL486 pKa = 3.91
MM1 pKa = 7.3HH2 pKa = 7.64PVFPGLSGALRR13 pKa = 11.84FGVHH17 pKa = 6.25NNNLRR22 pKa = 11.84NGCRR26 pKa = 11.84ALVEE30 pKa = 4.2RR31 pKa = 11.84VFLRR35 pKa = 11.84EE36 pKa = 4.14DD37 pKa = 3.43NGNLIPPPPCVEE49 pKa = 4.46DD50 pKa = 3.46VNKK53 pKa = 9.55TLEE56 pKa = 4.03EE57 pKa = 4.01FKK59 pKa = 10.86AAVITRR65 pKa = 11.84VGVHH69 pKa = 5.88RR70 pKa = 11.84RR71 pKa = 11.84WSVDD75 pKa = 2.8KK76 pKa = 10.69FVNSYY81 pKa = 8.12TGRR84 pKa = 11.84RR85 pKa = 11.84QNVYY89 pKa = 10.58ARR91 pKa = 11.84AAKK94 pKa = 10.23SLEE97 pKa = 4.06TTPVHH102 pKa = 6.0TGDD105 pKa = 3.97FKK107 pKa = 11.88VMNAFVKK114 pKa = 10.65AEE116 pKa = 4.07KK117 pKa = 10.13TNLTSKK123 pKa = 10.16PDD125 pKa = 3.68PAPRR129 pKa = 11.84VIQPRR134 pKa = 11.84NPRR137 pKa = 11.84YY138 pKa = 8.64NVEE141 pKa = 3.56VGRR144 pKa = 11.84FLKK147 pKa = 10.79GLEE150 pKa = 4.05HH151 pKa = 6.66KK152 pKa = 10.39VYY154 pKa = 10.61KK155 pKa = 10.4AVEE158 pKa = 4.73DD159 pKa = 3.63IFGDD163 pKa = 3.61PTVMKK168 pKa = 9.99GYY170 pKa = 9.64NAEE173 pKa = 3.99EE174 pKa = 4.42VAEE177 pKa = 4.17HH178 pKa = 6.42MVSKK182 pKa = 9.3WQSFSDD188 pKa = 4.06PVAVGLDD195 pKa = 3.17ASRR198 pKa = 11.84FDD200 pKa = 3.35QHH202 pKa = 6.42VRR204 pKa = 11.84PSMLEE209 pKa = 3.93WEE211 pKa = 4.36HH212 pKa = 7.3SVYY215 pKa = 10.78AGCFSTGEE223 pKa = 3.93AKK225 pKa = 10.4LLKK228 pKa = 9.68WLLQGQIRR236 pKa = 11.84NVCTMRR242 pKa = 11.84VTDD245 pKa = 4.03GLVKK249 pKa = 10.68YY250 pKa = 9.92RR251 pKa = 11.84VDD253 pKa = 3.56GSRR256 pKa = 11.84MSGDD260 pKa = 3.28MNTALGNCLIMCALVWRR277 pKa = 11.84LGKK280 pKa = 10.52VLGIRR285 pKa = 11.84LKK287 pKa = 10.72LANNGDD293 pKa = 3.83DD294 pKa = 3.31CVVFMEE300 pKa = 4.96RR301 pKa = 11.84KK302 pKa = 10.13DD303 pKa = 3.37LTAFNNKK310 pKa = 8.35VKK312 pKa = 10.61GFFLDD317 pKa = 3.29YY318 pKa = 10.67GFTMKK323 pKa = 10.45VEE325 pKa = 4.17APSFFIEE332 pKa = 4.95QIEE335 pKa = 4.55FCQAKK340 pKa = 9.42PVWDD344 pKa = 3.75GCKK347 pKa = 9.59WIMVRR352 pKa = 11.84DD353 pKa = 3.86PRR355 pKa = 11.84VCTSKK360 pKa = 11.04DD361 pKa = 3.14ATCVVKK367 pKa = 10.62DD368 pKa = 3.58YY369 pKa = 10.91GYY371 pKa = 11.48GSAASYY377 pKa = 9.86WLGAVGEE384 pKa = 4.99CGLAMAGGIPVMQNYY399 pKa = 9.36YY400 pKa = 10.61SAFARR405 pKa = 11.84FGNTNRR411 pKa = 11.84QVQCVTEE418 pKa = 3.85TGMAYY423 pKa = 10.65LARR426 pKa = 11.84GLHH429 pKa = 6.21RR430 pKa = 11.84EE431 pKa = 4.07ATVISADD438 pKa = 3.12ARR440 pKa = 11.84VSFWRR445 pKa = 11.84AFGIAPTQQRR455 pKa = 11.84EE456 pKa = 4.17LEE458 pKa = 4.23KK459 pKa = 10.48WCDD462 pKa = 3.57NTSLTLPNSPCIRR475 pKa = 11.84AYY477 pKa = 9.29PPRR480 pKa = 11.84GILPLLL486 pKa = 3.91
Molecular weight: 54.48 kDa
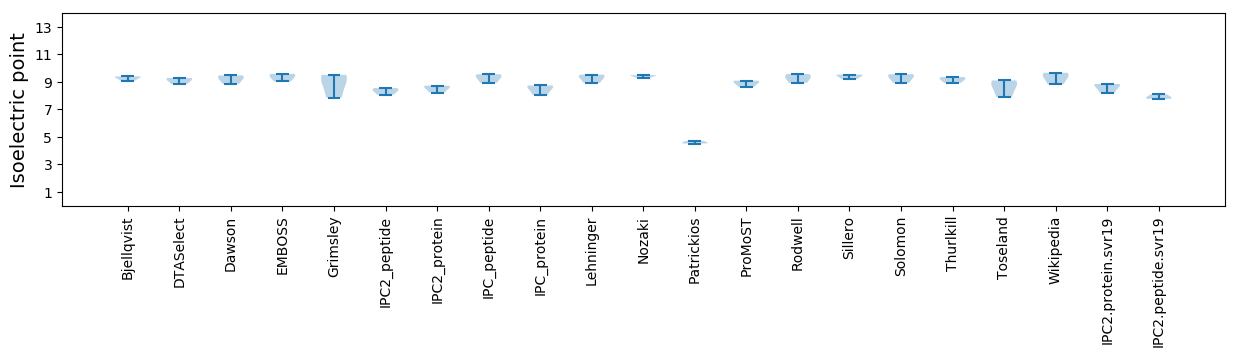
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG36|A0A1L3KG36_9VIRU RNA-directed RNA polymerase OS=Changjiang tombus-like virus 6 OX=1922820 PE=4 SV=1
MM1 pKa = 7.6AKK3 pKa = 10.07KK4 pKa = 10.35KK5 pKa = 10.12NSKK8 pKa = 9.75NKK10 pKa = 8.37TVIRR14 pKa = 11.84RR15 pKa = 11.84VAKK18 pKa = 10.27IPRR21 pKa = 11.84SPLDD25 pKa = 3.64AQGLAYY31 pKa = 10.18ARR33 pKa = 11.84LLADD37 pKa = 3.85PCNAPLVHH45 pKa = 6.36PVYY48 pKa = 10.83SGTEE52 pKa = 3.54GGYY55 pKa = 10.6LIRR58 pKa = 11.84ADD60 pKa = 3.46SFLTVGANALEE71 pKa = 4.17QAGVFQWVPGGIDD84 pKa = 3.28ALQDD88 pKa = 3.96NILLGAAANATTAINPGVSGAASPAWDD115 pKa = 4.03FLTNTASQVRR125 pKa = 11.84CVAACLRR132 pKa = 11.84IMYY135 pKa = 8.49PGSEE139 pKa = 4.16SSRR142 pKa = 11.84SGRR145 pKa = 11.84IAFGQASGALLKK157 pKa = 11.04SGVTVTANNLANTMAHH173 pKa = 5.83YY174 pKa = 10.69CRR176 pKa = 11.84TPADD180 pKa = 4.17EE181 pKa = 5.77IEE183 pKa = 4.84VVWKK187 pKa = 10.32PNDD190 pKa = 3.46ADD192 pKa = 3.57QLFRR196 pKa = 11.84NPALNGALQDD206 pKa = 3.82LDD208 pKa = 3.55RR209 pKa = 11.84RR210 pKa = 11.84AALVCSFAGLPAATGVVFRR229 pKa = 11.84MTAVYY234 pKa = 9.37EE235 pKa = 3.81WQPRR239 pKa = 11.84VNEE242 pKa = 4.95GIAVPNLSKK251 pKa = 11.17SMSSNTLDD259 pKa = 4.41DD260 pKa = 3.73VVNYY264 pKa = 9.72LIQRR268 pKa = 11.84GYY270 pKa = 11.58GFINGAAASAGHH282 pKa = 6.4GLSAGLISGISGVFGVMPSVTTSRR306 pKa = 11.84SRR308 pKa = 11.84RR309 pKa = 11.84VMGG312 pKa = 3.97
MM1 pKa = 7.6AKK3 pKa = 10.07KK4 pKa = 10.35KK5 pKa = 10.12NSKK8 pKa = 9.75NKK10 pKa = 8.37TVIRR14 pKa = 11.84RR15 pKa = 11.84VAKK18 pKa = 10.27IPRR21 pKa = 11.84SPLDD25 pKa = 3.64AQGLAYY31 pKa = 10.18ARR33 pKa = 11.84LLADD37 pKa = 3.85PCNAPLVHH45 pKa = 6.36PVYY48 pKa = 10.83SGTEE52 pKa = 3.54GGYY55 pKa = 10.6LIRR58 pKa = 11.84ADD60 pKa = 3.46SFLTVGANALEE71 pKa = 4.17QAGVFQWVPGGIDD84 pKa = 3.28ALQDD88 pKa = 3.96NILLGAAANATTAINPGVSGAASPAWDD115 pKa = 4.03FLTNTASQVRR125 pKa = 11.84CVAACLRR132 pKa = 11.84IMYY135 pKa = 8.49PGSEE139 pKa = 4.16SSRR142 pKa = 11.84SGRR145 pKa = 11.84IAFGQASGALLKK157 pKa = 11.04SGVTVTANNLANTMAHH173 pKa = 5.83YY174 pKa = 10.69CRR176 pKa = 11.84TPADD180 pKa = 4.17EE181 pKa = 5.77IEE183 pKa = 4.84VVWKK187 pKa = 10.32PNDD190 pKa = 3.46ADD192 pKa = 3.57QLFRR196 pKa = 11.84NPALNGALQDD206 pKa = 3.82LDD208 pKa = 3.55RR209 pKa = 11.84RR210 pKa = 11.84AALVCSFAGLPAATGVVFRR229 pKa = 11.84MTAVYY234 pKa = 9.37EE235 pKa = 3.81WQPRR239 pKa = 11.84VNEE242 pKa = 4.95GIAVPNLSKK251 pKa = 11.17SMSSNTLDD259 pKa = 4.41DD260 pKa = 3.73VVNYY264 pKa = 9.72LIQRR268 pKa = 11.84GYY270 pKa = 11.58GFINGAAASAGHH282 pKa = 6.4GLSAGLISGISGVFGVMPSVTTSRR306 pKa = 11.84SRR308 pKa = 11.84RR309 pKa = 11.84VMGG312 pKa = 3.97
Molecular weight: 32.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1064 |
266 |
486 |
354.7 |
38.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.842 ± 2.031 | 2.538 ± 0.331 |
4.605 ± 0.158 | 3.947 ± 0.651 |
4.041 ± 0.471 | 8.459 ± 0.403 |
1.504 ± 0.198 | 3.853 ± 0.396 |
4.135 ± 0.727 | 7.425 ± 0.669 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.289 ± 0.593 | 4.511 ± 1.21 |
4.793 ± 0.368 | 2.82 ± 0.134 |
7.331 ± 0.643 | 5.545 ± 0.872 |
5.169 ± 0.174 | 9.492 ± 0.409 |
1.692 ± 0.187 | 3.008 ± 0.174 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
