
Papaya lethal yellowing virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Sobemovirus
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
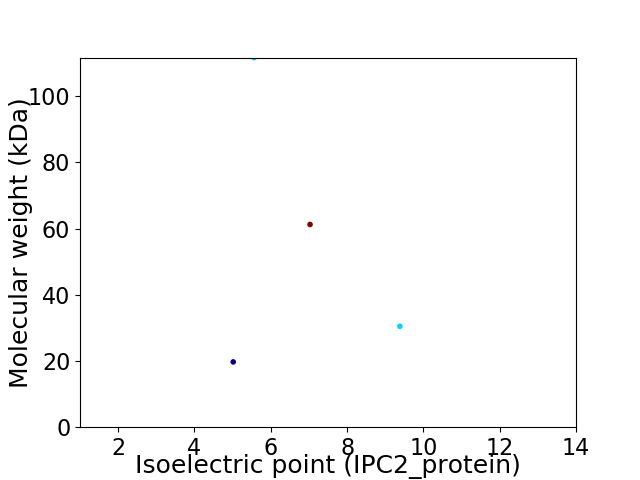
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J7G6A6|J7G6A6_9VIRU N-terminal protein OS=Papaya lethal yellowing virus OX=685899 GN=p2a PE=4 SV=1
MM1 pKa = 7.15SVVIEE6 pKa = 4.57VYY8 pKa = 10.66DD9 pKa = 3.76EE10 pKa = 4.14QSVSKK15 pKa = 10.72VLIEE19 pKa = 5.22SFKK22 pKa = 10.95LWPKK26 pKa = 10.79KK27 pKa = 10.76LGDD30 pKa = 4.48CVIQLKK36 pKa = 10.46GKK38 pKa = 8.48YY39 pKa = 9.36DD40 pKa = 3.44SSAVCRR46 pKa = 11.84GEE48 pKa = 3.89ITLCCEE54 pKa = 3.26KK55 pKa = 10.46CRR57 pKa = 11.84RR58 pKa = 11.84QEE60 pKa = 3.94IISFEE65 pKa = 4.12KK66 pKa = 10.42KK67 pKa = 10.09RR68 pKa = 11.84ILIHH72 pKa = 6.92CDD74 pKa = 2.83WEE76 pKa = 4.37NKK78 pKa = 9.9LCEE81 pKa = 4.74DD82 pKa = 3.65PCIDD86 pKa = 4.28RR87 pKa = 11.84FVKK90 pKa = 10.28PDD92 pKa = 3.2EE93 pKa = 5.08GFCNWCVEE101 pKa = 4.07DD102 pKa = 4.19HH103 pKa = 6.18QCTSEE108 pKa = 4.23GCSDD112 pKa = 4.3PDD114 pKa = 4.07CVFHH118 pKa = 7.69SDD120 pKa = 3.24SLEE123 pKa = 4.33FGWSEE128 pKa = 5.28SLNKK132 pKa = 9.79WIDD135 pKa = 3.28AHH137 pKa = 5.99KK138 pKa = 10.39HH139 pKa = 4.58GFVFDD144 pKa = 3.93RR145 pKa = 11.84YY146 pKa = 8.31EE147 pKa = 4.18AEE149 pKa = 3.96WYY151 pKa = 10.32HH152 pKa = 6.89PITAEE157 pKa = 3.84ILLSRR162 pKa = 11.84SPGLFRR168 pKa = 11.84DD169 pKa = 3.7
MM1 pKa = 7.15SVVIEE6 pKa = 4.57VYY8 pKa = 10.66DD9 pKa = 3.76EE10 pKa = 4.14QSVSKK15 pKa = 10.72VLIEE19 pKa = 5.22SFKK22 pKa = 10.95LWPKK26 pKa = 10.79KK27 pKa = 10.76LGDD30 pKa = 4.48CVIQLKK36 pKa = 10.46GKK38 pKa = 8.48YY39 pKa = 9.36DD40 pKa = 3.44SSAVCRR46 pKa = 11.84GEE48 pKa = 3.89ITLCCEE54 pKa = 3.26KK55 pKa = 10.46CRR57 pKa = 11.84RR58 pKa = 11.84QEE60 pKa = 3.94IISFEE65 pKa = 4.12KK66 pKa = 10.42KK67 pKa = 10.09RR68 pKa = 11.84ILIHH72 pKa = 6.92CDD74 pKa = 2.83WEE76 pKa = 4.37NKK78 pKa = 9.9LCEE81 pKa = 4.74DD82 pKa = 3.65PCIDD86 pKa = 4.28RR87 pKa = 11.84FVKK90 pKa = 10.28PDD92 pKa = 3.2EE93 pKa = 5.08GFCNWCVEE101 pKa = 4.07DD102 pKa = 4.19HH103 pKa = 6.18QCTSEE108 pKa = 4.23GCSDD112 pKa = 4.3PDD114 pKa = 4.07CVFHH118 pKa = 7.69SDD120 pKa = 3.24SLEE123 pKa = 4.33FGWSEE128 pKa = 5.28SLNKK132 pKa = 9.79WIDD135 pKa = 3.28AHH137 pKa = 5.99KK138 pKa = 10.39HH139 pKa = 4.58GFVFDD144 pKa = 3.93RR145 pKa = 11.84YY146 pKa = 8.31EE147 pKa = 4.18AEE149 pKa = 3.96WYY151 pKa = 10.32HH152 pKa = 6.89PITAEE157 pKa = 3.84ILLSRR162 pKa = 11.84SPGLFRR168 pKa = 11.84DD169 pKa = 3.7
Molecular weight: 19.72 kDa
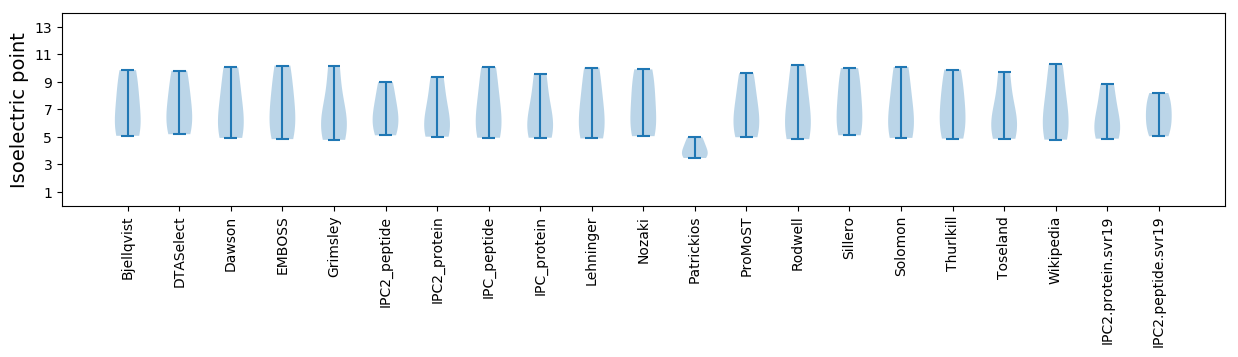
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J7GE58|J7GE58_9VIRU Capsid protein OS=Papaya lethal yellowing virus OX=685899 GN=cp PE=3 SV=1
MM1 pKa = 7.42ISAGRR6 pKa = 11.84VRR8 pKa = 11.84RR9 pKa = 11.84QTIKK13 pKa = 10.36NAARR17 pKa = 11.84AAAKK21 pKa = 10.33VAVQQAQRR29 pKa = 11.84VQQVAPKK36 pKa = 10.54GMNAKK41 pKa = 8.8QRR43 pKa = 11.84RR44 pKa = 11.84ALRR47 pKa = 11.84RR48 pKa = 11.84KK49 pKa = 8.29MLQSTASVQTRR60 pKa = 11.84QVTAPTAKK68 pKa = 9.38SVQVIRR74 pKa = 11.84AMPTLRR80 pKa = 11.84SRR82 pKa = 11.84NSVTGITHH90 pKa = 5.89MEE92 pKa = 3.94YY93 pKa = 10.32NQPVSVKK100 pKa = 8.83ATPTVTVDD108 pKa = 2.48NCMPYY113 pKa = 9.8TVGTWLRR120 pKa = 11.84GVASNWSKK128 pKa = 10.1WRR130 pKa = 11.84WRR132 pKa = 11.84KK133 pKa = 9.49LRR135 pKa = 11.84FYY137 pKa = 10.23YY138 pKa = 10.54LPSCATTTPGSVHH151 pKa = 6.27MGLTYY156 pKa = 10.52DD157 pKa = 4.26QYY159 pKa = 11.85DD160 pKa = 4.07TVPTNATEE168 pKa = 3.95MCNLQGYY175 pKa = 5.73TTGPVWSGAEE185 pKa = 3.84ASTALQNANVMCPPGAICCEE205 pKa = 3.73VDD207 pKa = 3.23VTRR210 pKa = 11.84FTQAWYY216 pKa = 10.13NYY218 pKa = 6.87ITSVSFTSATTVNVAIGNMYY238 pKa = 10.26SPVRR242 pKa = 11.84LVCLTDD248 pKa = 3.68EE249 pKa = 5.06GPATAVNAGKK259 pKa = 10.15ILVQYY264 pKa = 9.39EE265 pKa = 4.04IEE267 pKa = 4.61LIEE270 pKa = 4.45PVTSALNLL278 pKa = 3.6
MM1 pKa = 7.42ISAGRR6 pKa = 11.84VRR8 pKa = 11.84RR9 pKa = 11.84QTIKK13 pKa = 10.36NAARR17 pKa = 11.84AAAKK21 pKa = 10.33VAVQQAQRR29 pKa = 11.84VQQVAPKK36 pKa = 10.54GMNAKK41 pKa = 8.8QRR43 pKa = 11.84RR44 pKa = 11.84ALRR47 pKa = 11.84RR48 pKa = 11.84KK49 pKa = 8.29MLQSTASVQTRR60 pKa = 11.84QVTAPTAKK68 pKa = 9.38SVQVIRR74 pKa = 11.84AMPTLRR80 pKa = 11.84SRR82 pKa = 11.84NSVTGITHH90 pKa = 5.89MEE92 pKa = 3.94YY93 pKa = 10.32NQPVSVKK100 pKa = 8.83ATPTVTVDD108 pKa = 2.48NCMPYY113 pKa = 9.8TVGTWLRR120 pKa = 11.84GVASNWSKK128 pKa = 10.1WRR130 pKa = 11.84WRR132 pKa = 11.84KK133 pKa = 9.49LRR135 pKa = 11.84FYY137 pKa = 10.23YY138 pKa = 10.54LPSCATTTPGSVHH151 pKa = 6.27MGLTYY156 pKa = 10.52DD157 pKa = 4.26QYY159 pKa = 11.85DD160 pKa = 4.07TVPTNATEE168 pKa = 3.95MCNLQGYY175 pKa = 5.73TTGPVWSGAEE185 pKa = 3.84ASTALQNANVMCPPGAICCEE205 pKa = 3.73VDD207 pKa = 3.23VTRR210 pKa = 11.84FTQAWYY216 pKa = 10.13NYY218 pKa = 6.87ITSVSFTSATTVNVAIGNMYY238 pKa = 10.26SPVRR242 pKa = 11.84LVCLTDD248 pKa = 3.68EE249 pKa = 5.06GPATAVNAGKK259 pKa = 10.15ILVQYY264 pKa = 9.39EE265 pKa = 4.04IEE267 pKa = 4.61LIEE270 pKa = 4.45PVTSALNLL278 pKa = 3.6
Molecular weight: 30.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2009 |
169 |
1001 |
502.3 |
55.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.869 ± 1.042 | 3.285 ± 0.772 |
4.231 ± 0.751 | 6.969 ± 1.02 |
2.588 ± 0.558 | 7.566 ± 0.911 |
1.941 ± 0.34 | 4.729 ± 0.415 |
5.724 ± 0.626 | 8.86 ± 0.935 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.19 ± 0.353 | 4.032 ± 0.422 |
4.629 ± 0.417 | 2.787 ± 0.72 |
5.077 ± 0.501 | 8.213 ± 0.381 |
5.923 ± 1.272 | 8.213 ± 0.843 |
2.439 ± 0.227 | 3.733 ± 0.379 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
