
Ornithinibacillus sp. L9
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Ornithinibacillus
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
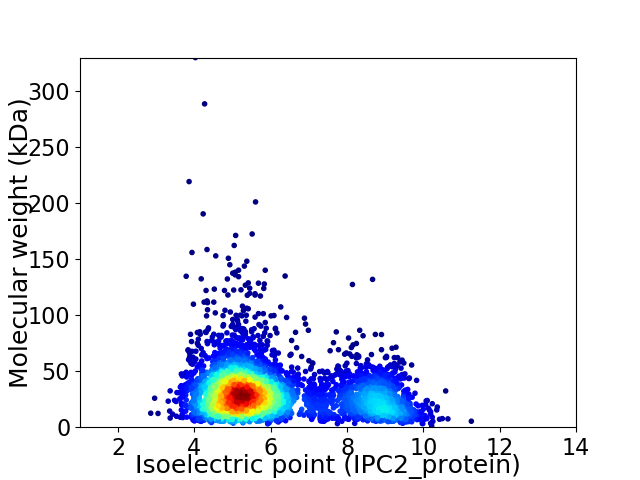
Virtual 2D-PAGE plot for 4042 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N8FHT5|A0A6N8FHT5_9BACI Uncharacterized protein OS=Ornithinibacillus sp. L9 OX=2678566 GN=GMD78_12280 PE=4 SV=1
MM1 pKa = 7.18NKK3 pKa = 9.86YY4 pKa = 9.92YY5 pKa = 10.47KK6 pKa = 10.4LAMIILFSGLLLIGCGSDD24 pKa = 4.81DD25 pKa = 5.37DD26 pKa = 4.93AAEE29 pKa = 4.35PNQGDD34 pKa = 4.28SYY36 pKa = 11.27PVNNKK41 pKa = 10.05DD42 pKa = 3.6EE43 pKa = 4.59SDD45 pKa = 3.82SNAEE49 pKa = 4.09NDD51 pKa = 3.79SEE53 pKa = 5.05SDD55 pKa = 3.54VSGSKK60 pKa = 8.27VTEE63 pKa = 3.5IHH65 pKa = 5.74TLEE68 pKa = 4.49EE69 pKa = 4.01EE70 pKa = 4.55DD71 pKa = 5.72DD72 pKa = 3.53ITRR75 pKa = 11.84EE76 pKa = 4.11NISINYY82 pKa = 8.72DD83 pKa = 3.15GSVVYY88 pKa = 10.24FDD90 pKa = 5.52LVDD93 pKa = 4.63DD94 pKa = 4.42SGEE97 pKa = 4.41DD98 pKa = 3.67EE99 pKa = 4.49TNTPYY104 pKa = 10.42FYY106 pKa = 11.32YY107 pKa = 10.72NGDD110 pKa = 3.54MFDD113 pKa = 3.75ASSVDD118 pKa = 3.46SFSDD122 pKa = 3.16CKK124 pKa = 11.33YY125 pKa = 10.65NVNAADD131 pKa = 4.04TIYY134 pKa = 9.3FTGTCKK140 pKa = 10.5DD141 pKa = 3.56EE142 pKa = 4.47KK143 pKa = 9.65DD144 pKa = 3.46TRR146 pKa = 11.84VSVVYY151 pKa = 10.51DD152 pKa = 3.1ITEE155 pKa = 3.85NTITHH160 pKa = 5.23STDD163 pKa = 2.93RR164 pKa = 11.84DD165 pKa = 3.82RR166 pKa = 11.84NVHH169 pKa = 5.13VLNDD173 pKa = 3.34GRR175 pKa = 11.84VLYY178 pKa = 8.11TTEE181 pKa = 3.99FSEE184 pKa = 5.67DD185 pKa = 3.02GTIYY189 pKa = 10.8EE190 pKa = 4.12LTEE193 pKa = 4.71DD194 pKa = 3.89GEE196 pKa = 4.65EE197 pKa = 3.95PFIEE201 pKa = 5.3IEE203 pKa = 4.76DD204 pKa = 3.86MPTILNFSVDD214 pKa = 3.47QNNEE218 pKa = 3.35VFFIYY223 pKa = 10.09GANDD227 pKa = 3.9EE228 pKa = 4.48YY229 pKa = 10.66WNSYY233 pKa = 9.5VFDD236 pKa = 3.91TTSDD240 pKa = 3.74SEE242 pKa = 4.31PVPFQSVPDD251 pKa = 4.3DD252 pKa = 4.63DD253 pKa = 4.57EE254 pKa = 4.37ATTVEE259 pKa = 4.63GRR261 pKa = 11.84ISPDD265 pKa = 2.45GKK267 pKa = 10.17YY268 pKa = 10.49IMYY271 pKa = 9.67RR272 pKa = 11.84YY273 pKa = 10.0RR274 pKa = 11.84GVTGGKK280 pKa = 9.15HH281 pKa = 4.08YY282 pKa = 11.02NYY284 pKa = 9.99VDD286 pKa = 3.31SYY288 pKa = 11.16IYY290 pKa = 10.82NRR292 pKa = 11.84DD293 pKa = 3.51SEE295 pKa = 4.68EE296 pKa = 4.25EE297 pKa = 3.63IHH299 pKa = 6.65LGHH302 pKa = 6.55GFDD305 pKa = 3.84MNFVRR310 pKa = 11.84SNGFVFDD317 pKa = 3.73EE318 pKa = 4.07VRR320 pKa = 11.84EE321 pKa = 4.06YY322 pKa = 11.44GQVIYY327 pKa = 11.25NMEE330 pKa = 4.37TDD332 pKa = 2.93TWLAPDD338 pKa = 4.16TADD341 pKa = 2.71ITYY344 pKa = 10.26FDD346 pKa = 3.54STYY349 pKa = 10.66KK350 pKa = 10.72DD351 pKa = 3.55EE352 pKa = 4.93YY353 pKa = 10.95LDD355 pKa = 4.3EE356 pKa = 5.89ADD358 pKa = 4.31QEE360 pKa = 4.37DD361 pKa = 4.3FQRR364 pKa = 11.84SSFVAISGDD373 pKa = 3.54GEE375 pKa = 4.66TVLTLDD381 pKa = 3.83RR382 pKa = 11.84FRR384 pKa = 11.84PLSEE388 pKa = 4.65DD389 pKa = 2.89RR390 pKa = 11.84VEE392 pKa = 3.89YY393 pKa = 10.93SKK395 pKa = 11.38LHH397 pKa = 5.98TISTEE402 pKa = 3.95DD403 pKa = 3.45YY404 pKa = 9.86IKK406 pKa = 10.64FLEE409 pKa = 4.82EE410 pKa = 3.6YY411 pKa = 10.21DD412 pKa = 3.65IEE414 pKa = 5.0IDD416 pKa = 4.18FEE418 pKa = 4.37PSPFDD423 pKa = 3.5EE424 pKa = 5.29KK425 pKa = 11.35GAA427 pKa = 3.85
MM1 pKa = 7.18NKK3 pKa = 9.86YY4 pKa = 9.92YY5 pKa = 10.47KK6 pKa = 10.4LAMIILFSGLLLIGCGSDD24 pKa = 4.81DD25 pKa = 5.37DD26 pKa = 4.93AAEE29 pKa = 4.35PNQGDD34 pKa = 4.28SYY36 pKa = 11.27PVNNKK41 pKa = 10.05DD42 pKa = 3.6EE43 pKa = 4.59SDD45 pKa = 3.82SNAEE49 pKa = 4.09NDD51 pKa = 3.79SEE53 pKa = 5.05SDD55 pKa = 3.54VSGSKK60 pKa = 8.27VTEE63 pKa = 3.5IHH65 pKa = 5.74TLEE68 pKa = 4.49EE69 pKa = 4.01EE70 pKa = 4.55DD71 pKa = 5.72DD72 pKa = 3.53ITRR75 pKa = 11.84EE76 pKa = 4.11NISINYY82 pKa = 8.72DD83 pKa = 3.15GSVVYY88 pKa = 10.24FDD90 pKa = 5.52LVDD93 pKa = 4.63DD94 pKa = 4.42SGEE97 pKa = 4.41DD98 pKa = 3.67EE99 pKa = 4.49TNTPYY104 pKa = 10.42FYY106 pKa = 11.32YY107 pKa = 10.72NGDD110 pKa = 3.54MFDD113 pKa = 3.75ASSVDD118 pKa = 3.46SFSDD122 pKa = 3.16CKK124 pKa = 11.33YY125 pKa = 10.65NVNAADD131 pKa = 4.04TIYY134 pKa = 9.3FTGTCKK140 pKa = 10.5DD141 pKa = 3.56EE142 pKa = 4.47KK143 pKa = 9.65DD144 pKa = 3.46TRR146 pKa = 11.84VSVVYY151 pKa = 10.51DD152 pKa = 3.1ITEE155 pKa = 3.85NTITHH160 pKa = 5.23STDD163 pKa = 2.93RR164 pKa = 11.84DD165 pKa = 3.82RR166 pKa = 11.84NVHH169 pKa = 5.13VLNDD173 pKa = 3.34GRR175 pKa = 11.84VLYY178 pKa = 8.11TTEE181 pKa = 3.99FSEE184 pKa = 5.67DD185 pKa = 3.02GTIYY189 pKa = 10.8EE190 pKa = 4.12LTEE193 pKa = 4.71DD194 pKa = 3.89GEE196 pKa = 4.65EE197 pKa = 3.95PFIEE201 pKa = 5.3IEE203 pKa = 4.76DD204 pKa = 3.86MPTILNFSVDD214 pKa = 3.47QNNEE218 pKa = 3.35VFFIYY223 pKa = 10.09GANDD227 pKa = 3.9EE228 pKa = 4.48YY229 pKa = 10.66WNSYY233 pKa = 9.5VFDD236 pKa = 3.91TTSDD240 pKa = 3.74SEE242 pKa = 4.31PVPFQSVPDD251 pKa = 4.3DD252 pKa = 4.63DD253 pKa = 4.57EE254 pKa = 4.37ATTVEE259 pKa = 4.63GRR261 pKa = 11.84ISPDD265 pKa = 2.45GKK267 pKa = 10.17YY268 pKa = 10.49IMYY271 pKa = 9.67RR272 pKa = 11.84YY273 pKa = 10.0RR274 pKa = 11.84GVTGGKK280 pKa = 9.15HH281 pKa = 4.08YY282 pKa = 11.02NYY284 pKa = 9.99VDD286 pKa = 3.31SYY288 pKa = 11.16IYY290 pKa = 10.82NRR292 pKa = 11.84DD293 pKa = 3.51SEE295 pKa = 4.68EE296 pKa = 4.25EE297 pKa = 3.63IHH299 pKa = 6.65LGHH302 pKa = 6.55GFDD305 pKa = 3.84MNFVRR310 pKa = 11.84SNGFVFDD317 pKa = 3.73EE318 pKa = 4.07VRR320 pKa = 11.84EE321 pKa = 4.06YY322 pKa = 11.44GQVIYY327 pKa = 11.25NMEE330 pKa = 4.37TDD332 pKa = 2.93TWLAPDD338 pKa = 4.16TADD341 pKa = 2.71ITYY344 pKa = 10.26FDD346 pKa = 3.54STYY349 pKa = 10.66KK350 pKa = 10.72DD351 pKa = 3.55EE352 pKa = 4.93YY353 pKa = 10.95LDD355 pKa = 4.3EE356 pKa = 5.89ADD358 pKa = 4.31QEE360 pKa = 4.37DD361 pKa = 4.3FQRR364 pKa = 11.84SSFVAISGDD373 pKa = 3.54GEE375 pKa = 4.66TVLTLDD381 pKa = 3.83RR382 pKa = 11.84FRR384 pKa = 11.84PLSEE388 pKa = 4.65DD389 pKa = 2.89RR390 pKa = 11.84VEE392 pKa = 3.89YY393 pKa = 10.93SKK395 pKa = 11.38LHH397 pKa = 5.98TISTEE402 pKa = 3.95DD403 pKa = 3.45YY404 pKa = 9.86IKK406 pKa = 10.64FLEE409 pKa = 4.82EE410 pKa = 3.6YY411 pKa = 10.21DD412 pKa = 3.65IEE414 pKa = 5.0IDD416 pKa = 4.18FEE418 pKa = 4.37PSPFDD423 pKa = 3.5EE424 pKa = 5.29KK425 pKa = 11.35GAA427 pKa = 3.85
Molecular weight: 48.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N8FEN4|A0A6N8FEN4_9BACI Hemolysin III family protein OS=Ornithinibacillus sp. L9 OX=2678566 GN=GMD78_06170 PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.12KK14 pKa = 8.46VHH16 pKa = 5.5GFRR19 pKa = 11.84TRR21 pKa = 11.84MSTKK25 pKa = 10.06NGRR28 pKa = 11.84KK29 pKa = 8.49VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.12KK14 pKa = 8.46VHH16 pKa = 5.5GFRR19 pKa = 11.84TRR21 pKa = 11.84MSTKK25 pKa = 10.06NGRR28 pKa = 11.84KK29 pKa = 8.49VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
Molecular weight: 5.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1157730 |
10 |
3029 |
286.4 |
32.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.318 ± 0.039 | 0.623 ± 0.01 |
5.482 ± 0.034 | 7.813 ± 0.05 |
4.547 ± 0.031 | 6.66 ± 0.041 |
2.147 ± 0.021 | 8.302 ± 0.041 |
6.653 ± 0.041 | 9.503 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.746 ± 0.019 | 4.822 ± 0.03 |
3.52 ± 0.022 | 3.737 ± 0.024 |
3.865 ± 0.03 | 5.994 ± 0.031 |
5.481 ± 0.029 | 7.027 ± 0.034 |
1.043 ± 0.017 | 3.716 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
