
Hubei tetragnatha maxillosa virus 6
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.16
Get precalculated fractions of proteins
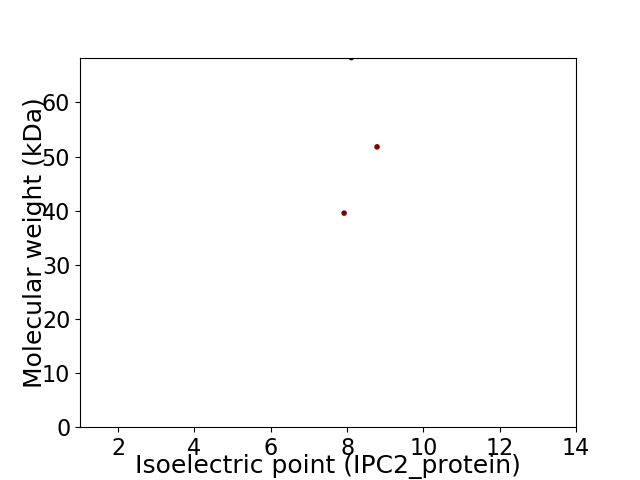
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEQ2|A0A1L3KEQ2_9VIRU RNA-directed RNA polymerase OS=Hubei tetragnatha maxillosa virus 6 OX=1923248 PE=4 SV=1
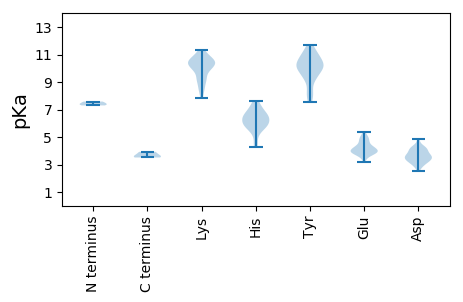
MM1 pKa = 7.3TSSPGYY7 pKa = 9.29PYY9 pKa = 10.03IYY11 pKa = 9.84HH12 pKa = 6.24HH13 pKa = 6.92TNNASFFHH21 pKa = 6.42YY22 pKa = 10.12KK23 pKa = 10.57DD24 pKa = 3.56GVVDD28 pKa = 4.17PDD30 pKa = 3.33RR31 pKa = 11.84LKK33 pKa = 11.11VVWEE37 pKa = 3.82MVINQIQEE45 pKa = 3.95RR46 pKa = 11.84RR47 pKa = 11.84SDD49 pKa = 4.1PIRR52 pKa = 11.84IFVKK56 pKa = 10.43PEE58 pKa = 3.16PHH60 pKa = 6.69SYY62 pKa = 10.78KK63 pKa = 10.44KK64 pKa = 10.4LQSGRR69 pKa = 11.84YY70 pKa = 8.26RR71 pKa = 11.84LISSVSIIDD80 pKa = 3.78QIIDD84 pKa = 3.08AMLFRR89 pKa = 11.84EE90 pKa = 4.81MNAQMIAQWPLLPTRR105 pKa = 11.84VGWSPYY111 pKa = 8.19YY112 pKa = 10.55GGWKK116 pKa = 8.5MMPYY120 pKa = 9.96VRR122 pKa = 11.84EE123 pKa = 3.51QMAIDD128 pKa = 3.73KK129 pKa = 10.71SSWDD133 pKa = 3.24WTVQPWIVEE142 pKa = 3.84AVLQIRR148 pKa = 11.84TEE150 pKa = 3.92MCANLTPQWEE160 pKa = 4.25EE161 pKa = 3.58LARR164 pKa = 11.84WRR166 pKa = 11.84YY167 pKa = 9.29KK168 pKa = 10.41EE169 pKa = 5.17LYY171 pKa = 9.99GSPLFVTTRR180 pKa = 11.84GSIFKK185 pKa = 9.91QQTPGVMKK193 pKa = 10.31SGCYY197 pKa = 7.69NTLMDD202 pKa = 4.32NSIMQLILHH211 pKa = 6.64ALVTQKK217 pKa = 11.29LNIPFGQIIAMGDD230 pKa = 3.46DD231 pKa = 3.85TLQDD235 pKa = 4.05ALPEE239 pKa = 5.04PILTDD244 pKa = 3.62YY245 pKa = 11.39LDD247 pKa = 3.8QLSMYY252 pKa = 10.07CNVKK256 pKa = 9.96LCEE259 pKa = 4.34KK260 pKa = 10.31KK261 pKa = 10.35RR262 pKa = 11.84EE263 pKa = 3.92FCGFNFNVNGVVEE276 pKa = 4.45PIYY279 pKa = 10.23RR280 pKa = 11.84GKK282 pKa = 9.81HH283 pKa = 4.67AYY285 pKa = 9.43KK286 pKa = 9.09MLHH289 pKa = 5.6MPIDD293 pKa = 4.01IMQSMADD300 pKa = 4.13SYY302 pKa = 11.63TMMYY306 pKa = 9.56HH307 pKa = 7.1KK308 pKa = 10.39SRR310 pKa = 11.84YY311 pKa = 9.22SPLIDD316 pKa = 3.15RR317 pKa = 11.84FFRR320 pKa = 11.84KK321 pKa = 9.57IGCEE325 pKa = 3.83VSPRR329 pKa = 11.84PYY331 pKa = 10.21RR332 pKa = 11.84DD333 pKa = 2.94LVYY336 pKa = 11.05GG337 pKa = 3.92
MM1 pKa = 7.3TSSPGYY7 pKa = 9.29PYY9 pKa = 10.03IYY11 pKa = 9.84HH12 pKa = 6.24HH13 pKa = 6.92TNNASFFHH21 pKa = 6.42YY22 pKa = 10.12KK23 pKa = 10.57DD24 pKa = 3.56GVVDD28 pKa = 4.17PDD30 pKa = 3.33RR31 pKa = 11.84LKK33 pKa = 11.11VVWEE37 pKa = 3.82MVINQIQEE45 pKa = 3.95RR46 pKa = 11.84RR47 pKa = 11.84SDD49 pKa = 4.1PIRR52 pKa = 11.84IFVKK56 pKa = 10.43PEE58 pKa = 3.16PHH60 pKa = 6.69SYY62 pKa = 10.78KK63 pKa = 10.44KK64 pKa = 10.4LQSGRR69 pKa = 11.84YY70 pKa = 8.26RR71 pKa = 11.84LISSVSIIDD80 pKa = 3.78QIIDD84 pKa = 3.08AMLFRR89 pKa = 11.84EE90 pKa = 4.81MNAQMIAQWPLLPTRR105 pKa = 11.84VGWSPYY111 pKa = 8.19YY112 pKa = 10.55GGWKK116 pKa = 8.5MMPYY120 pKa = 9.96VRR122 pKa = 11.84EE123 pKa = 3.51QMAIDD128 pKa = 3.73KK129 pKa = 10.71SSWDD133 pKa = 3.24WTVQPWIVEE142 pKa = 3.84AVLQIRR148 pKa = 11.84TEE150 pKa = 3.92MCANLTPQWEE160 pKa = 4.25EE161 pKa = 3.58LARR164 pKa = 11.84WRR166 pKa = 11.84YY167 pKa = 9.29KK168 pKa = 10.41EE169 pKa = 5.17LYY171 pKa = 9.99GSPLFVTTRR180 pKa = 11.84GSIFKK185 pKa = 9.91QQTPGVMKK193 pKa = 10.31SGCYY197 pKa = 7.69NTLMDD202 pKa = 4.32NSIMQLILHH211 pKa = 6.64ALVTQKK217 pKa = 11.29LNIPFGQIIAMGDD230 pKa = 3.46DD231 pKa = 3.85TLQDD235 pKa = 4.05ALPEE239 pKa = 5.04PILTDD244 pKa = 3.62YY245 pKa = 11.39LDD247 pKa = 3.8QLSMYY252 pKa = 10.07CNVKK256 pKa = 9.96LCEE259 pKa = 4.34KK260 pKa = 10.31KK261 pKa = 10.35RR262 pKa = 11.84EE263 pKa = 3.92FCGFNFNVNGVVEE276 pKa = 4.45PIYY279 pKa = 10.23RR280 pKa = 11.84GKK282 pKa = 9.81HH283 pKa = 4.67AYY285 pKa = 9.43KK286 pKa = 9.09MLHH289 pKa = 5.6MPIDD293 pKa = 4.01IMQSMADD300 pKa = 4.13SYY302 pKa = 11.63TMMYY306 pKa = 9.56HH307 pKa = 7.1KK308 pKa = 10.39SRR310 pKa = 11.84YY311 pKa = 9.22SPLIDD316 pKa = 3.15RR317 pKa = 11.84FFRR320 pKa = 11.84KK321 pKa = 9.57IGCEE325 pKa = 3.83VSPRR329 pKa = 11.84PYY331 pKa = 10.21RR332 pKa = 11.84DD333 pKa = 2.94LVYY336 pKa = 11.05GG337 pKa = 3.92
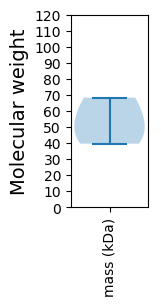
Molecular weight: 39.56 kDa
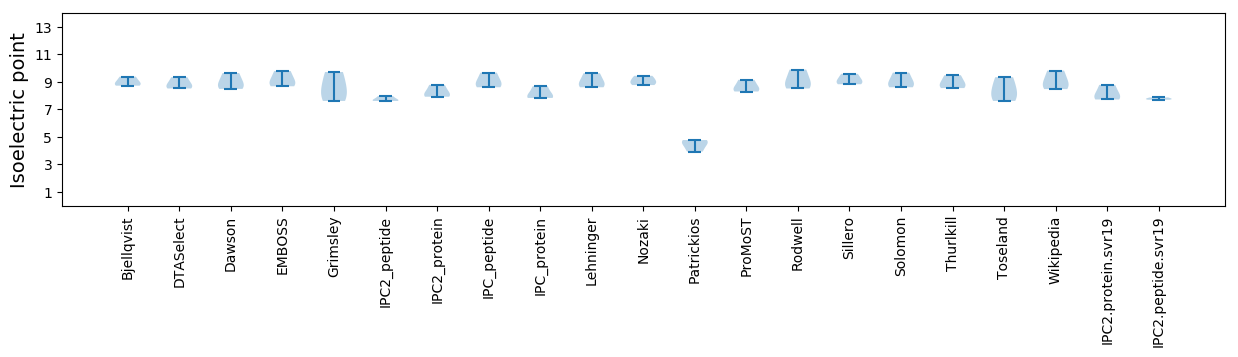
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEN8|A0A1L3KEN8_9VIRU C2H2-type domain-containing protein OS=Hubei tetragnatha maxillosa virus 6 OX=1923248 PE=4 SV=1
MM1 pKa = 7.58SSNKK5 pKa = 9.13TFVCPRR11 pKa = 11.84DD12 pKa = 3.3SRR14 pKa = 11.84KK15 pKa = 9.87FKK17 pKa = 10.29TRR19 pKa = 11.84AALQQHH25 pKa = 6.19MAASHH30 pKa = 5.19PQGRR34 pKa = 11.84PSPAPKK40 pKa = 10.14APRR43 pKa = 11.84RR44 pKa = 11.84QGNNTRR50 pKa = 11.84VPIVVPGSNGAFVRR64 pKa = 11.84FRR66 pKa = 11.84NKK68 pKa = 10.12EE69 pKa = 3.85FLKK72 pKa = 10.68EE73 pKa = 3.74LSFPTKK79 pKa = 9.68GTTLHH84 pKa = 6.24VGVHH88 pKa = 5.72FNIAGDD94 pKa = 3.86APILKK99 pKa = 9.75KK100 pKa = 10.19MSSIYY105 pKa = 10.33DD106 pKa = 3.35SYY108 pKa = 10.96RR109 pKa = 11.84IHH111 pKa = 6.02GVKK114 pKa = 10.3YY115 pKa = 10.11HH116 pKa = 5.92FTTSVSKK123 pKa = 7.86TTSGLVAMAVDD134 pKa = 4.34PGVTKK139 pKa = 11.12YY140 pKa = 10.68PGDD143 pKa = 3.65LKK145 pKa = 10.85ATLSANPHH153 pKa = 4.33VTGPIHH159 pKa = 7.58SDD161 pKa = 2.78RR162 pKa = 11.84LSITVPTVWCNPMLLRR178 pKa = 11.84EE179 pKa = 4.34VGSSNATPFQLIASVKK195 pKa = 9.31MSADD199 pKa = 3.39QPSNTIVGYY208 pKa = 10.13IEE210 pKa = 3.91IEE212 pKa = 3.81YY213 pKa = 10.31DD214 pKa = 3.21IEE216 pKa = 4.97LSGLTPXAEE225 pKa = 3.98QAGGVVAEE233 pKa = 4.07QHH235 pKa = 6.08NFTEE239 pKa = 4.82GAAARR244 pKa = 11.84IEE246 pKa = 4.12YY247 pKa = 9.98RR248 pKa = 11.84SQAKK252 pKa = 9.27EE253 pKa = 3.63PNYY256 pKa = 10.14YY257 pKa = 10.78ASNLLTNMPTAQYY270 pKa = 11.15LKK272 pKa = 11.24GEE274 pKa = 4.32LDD276 pKa = 3.49YY277 pKa = 11.66DD278 pKa = 4.03FTKK281 pKa = 10.05STTPPSVLGPVEE293 pKa = 4.78EE294 pKa = 4.62INSTGIPQWNVDD306 pKa = 4.24SYY308 pKa = 11.51PHH310 pKa = 6.42TNLTEE315 pKa = 4.97DD316 pKa = 3.52INVQTRR322 pKa = 11.84IVGSMQMTGILFVNWVSFIDD342 pKa = 4.33FLGLDD347 pKa = 3.66VEE349 pKa = 4.69STKK352 pKa = 11.0GFILTASSTVCDD364 pKa = 4.34VIFNAQPKK372 pKa = 9.38IVWNAAKK379 pKa = 8.54STCYY383 pKa = 10.59ASVGMWLKK391 pKa = 9.83TKK393 pKa = 10.21TEE395 pKa = 3.97APVFPVLTLEE405 pKa = 4.79LDD407 pKa = 4.89FIGHH411 pKa = 5.8RR412 pKa = 11.84HH413 pKa = 5.45SGEE416 pKa = 3.61NAIFFVYY423 pKa = 9.57HH424 pKa = 5.84HH425 pKa = 6.76VSMAFSKK432 pKa = 10.71LKK434 pKa = 10.23SLPWSMDD441 pKa = 2.82YY442 pKa = 10.87DD443 pKa = 3.99VPRR446 pKa = 11.84KK447 pKa = 8.56PKK449 pKa = 7.98YY450 pKa = 9.0WFVRR454 pKa = 11.84KK455 pKa = 8.47YY456 pKa = 7.58WRR458 pKa = 11.84KK459 pKa = 9.2FKK461 pKa = 10.6TSAKK465 pKa = 10.16RR466 pKa = 11.84NN467 pKa = 3.61
MM1 pKa = 7.58SSNKK5 pKa = 9.13TFVCPRR11 pKa = 11.84DD12 pKa = 3.3SRR14 pKa = 11.84KK15 pKa = 9.87FKK17 pKa = 10.29TRR19 pKa = 11.84AALQQHH25 pKa = 6.19MAASHH30 pKa = 5.19PQGRR34 pKa = 11.84PSPAPKK40 pKa = 10.14APRR43 pKa = 11.84RR44 pKa = 11.84QGNNTRR50 pKa = 11.84VPIVVPGSNGAFVRR64 pKa = 11.84FRR66 pKa = 11.84NKK68 pKa = 10.12EE69 pKa = 3.85FLKK72 pKa = 10.68EE73 pKa = 3.74LSFPTKK79 pKa = 9.68GTTLHH84 pKa = 6.24VGVHH88 pKa = 5.72FNIAGDD94 pKa = 3.86APILKK99 pKa = 9.75KK100 pKa = 10.19MSSIYY105 pKa = 10.33DD106 pKa = 3.35SYY108 pKa = 10.96RR109 pKa = 11.84IHH111 pKa = 6.02GVKK114 pKa = 10.3YY115 pKa = 10.11HH116 pKa = 5.92FTTSVSKK123 pKa = 7.86TTSGLVAMAVDD134 pKa = 4.34PGVTKK139 pKa = 11.12YY140 pKa = 10.68PGDD143 pKa = 3.65LKK145 pKa = 10.85ATLSANPHH153 pKa = 4.33VTGPIHH159 pKa = 7.58SDD161 pKa = 2.78RR162 pKa = 11.84LSITVPTVWCNPMLLRR178 pKa = 11.84EE179 pKa = 4.34VGSSNATPFQLIASVKK195 pKa = 9.31MSADD199 pKa = 3.39QPSNTIVGYY208 pKa = 10.13IEE210 pKa = 3.91IEE212 pKa = 3.81YY213 pKa = 10.31DD214 pKa = 3.21IEE216 pKa = 4.97LSGLTPXAEE225 pKa = 3.98QAGGVVAEE233 pKa = 4.07QHH235 pKa = 6.08NFTEE239 pKa = 4.82GAAARR244 pKa = 11.84IEE246 pKa = 4.12YY247 pKa = 9.98RR248 pKa = 11.84SQAKK252 pKa = 9.27EE253 pKa = 3.63PNYY256 pKa = 10.14YY257 pKa = 10.78ASNLLTNMPTAQYY270 pKa = 11.15LKK272 pKa = 11.24GEE274 pKa = 4.32LDD276 pKa = 3.49YY277 pKa = 11.66DD278 pKa = 4.03FTKK281 pKa = 10.05STTPPSVLGPVEE293 pKa = 4.78EE294 pKa = 4.62INSTGIPQWNVDD306 pKa = 4.24SYY308 pKa = 11.51PHH310 pKa = 6.42TNLTEE315 pKa = 4.97DD316 pKa = 3.52INVQTRR322 pKa = 11.84IVGSMQMTGILFVNWVSFIDD342 pKa = 4.33FLGLDD347 pKa = 3.66VEE349 pKa = 4.69STKK352 pKa = 11.0GFILTASSTVCDD364 pKa = 4.34VIFNAQPKK372 pKa = 9.38IVWNAAKK379 pKa = 8.54STCYY383 pKa = 10.59ASVGMWLKK391 pKa = 9.83TKK393 pKa = 10.21TEE395 pKa = 3.97APVFPVLTLEE405 pKa = 4.79LDD407 pKa = 4.89FIGHH411 pKa = 5.8RR412 pKa = 11.84HH413 pKa = 5.45SGEE416 pKa = 3.61NAIFFVYY423 pKa = 9.57HH424 pKa = 5.84HH425 pKa = 6.76VSMAFSKK432 pKa = 10.71LKK434 pKa = 10.23SLPWSMDD441 pKa = 2.82YY442 pKa = 10.87DD443 pKa = 3.99VPRR446 pKa = 11.84KK447 pKa = 8.56PKK449 pKa = 7.98YY450 pKa = 9.0WFVRR454 pKa = 11.84KK455 pKa = 8.47YY456 pKa = 7.58WRR458 pKa = 11.84KK459 pKa = 9.2FKK461 pKa = 10.6TSAKK465 pKa = 10.16RR466 pKa = 11.84NN467 pKa = 3.61
Molecular weight: 51.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1405 |
337 |
601 |
468.3 |
53.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.623 ± 0.762 | 1.352 ± 0.214 |
4.27 ± 0.352 | 5.836 ± 1.075 |
4.057 ± 0.357 | 5.907 ± 0.293 |
3.06 ± 0.255 | 6.263 ± 0.563 |
6.69 ± 0.535 | 7.046 ± 0.257 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.915 ± 0.735 | 3.488 ± 0.65 |
6.121 ± 0.403 | 3.701 ± 0.544 |
5.409 ± 0.463 | 7.544 ± 0.528 |
6.762 ± 0.868 | 6.975 ± 0.481 |
1.566 ± 0.421 | 4.342 ± 0.621 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
