
Zoogloea oleivorans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Rhodocyclales; Zoogloeaceae; Zoogloea
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
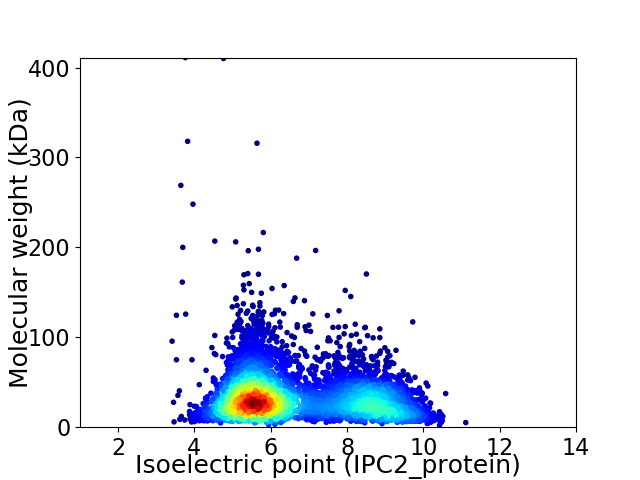
Virtual 2D-PAGE plot for 4972 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6C2D181|A0A6C2D181_9RHOO 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase OS=Zoogloea oleivorans OX=1552750 GN=uraD PE=4 SV=1
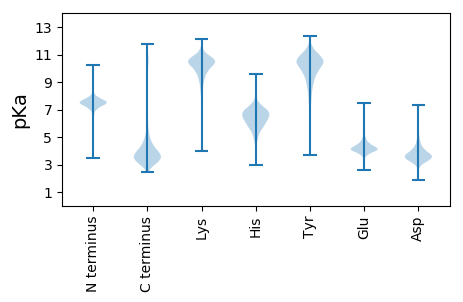
MM1 pKa = 7.58SGGGDD6 pKa = 3.3VTRR9 pKa = 11.84FTIDD13 pKa = 3.39PDD15 pKa = 3.49TGALSFLTAPDD26 pKa = 3.86FEE28 pKa = 4.7VPQDD32 pKa = 3.27EE33 pKa = 4.83GANNVYY39 pKa = 10.6DD40 pKa = 4.36VIVQVMDD47 pKa = 4.18GANSTAIQAITVTVTAVNDD66 pKa = 3.6NAPVLTSNGGGVMAAVTIAEE86 pKa = 4.41STTLVTTLTATDD98 pKa = 3.7SDD100 pKa = 4.53LPGDD104 pKa = 3.66TLYY107 pKa = 11.42YY108 pKa = 10.34SIVGGADD115 pKa = 2.98AAKK118 pKa = 10.0FAIDD122 pKa = 3.79TFSGALNFALAPDD135 pKa = 3.96YY136 pKa = 10.86DD137 pKa = 3.97LPGDD141 pKa = 3.65VGGDD145 pKa = 3.46NIYY148 pKa = 11.15DD149 pKa = 3.69VIVRR153 pKa = 11.84VDD155 pKa = 3.45DD156 pKa = 3.35GTYY159 pKa = 10.97AVNQSLAITVSNTNEE174 pKa = 3.89APAVTITSTSYY185 pKa = 9.72TATEE189 pKa = 3.81QSALVLGGTGLSISDD204 pKa = 4.08ADD206 pKa = 3.86AGSHH210 pKa = 5.08VVSARR215 pKa = 11.84LNVVSGTLSATAGSSGVTITGSGTASLVFDD245 pKa = 3.97GTLAQINDD253 pKa = 3.78FLAGKK258 pKa = 9.94LGASLTYY265 pKa = 10.55LIDD268 pKa = 4.02IDD270 pKa = 4.49TPPASDD276 pKa = 3.86TLTLAADD283 pKa = 4.26DD284 pKa = 4.9LGHH287 pKa = 6.6TGSGTAASGSKK298 pKa = 10.23SVLINITAVNDD309 pKa = 4.13GPANTTPGTQVTLVNTPLVFGVSHH333 pKa = 6.63GNALSVTDD341 pKa = 4.51SDD343 pKa = 4.42AAANPLQISLVATHH357 pKa = 6.43GTLTLGNTTGLTFVSGDD374 pKa = 3.37GAADD378 pKa = 3.25AGMTFTGTQASINAALEE395 pKa = 4.28GLSYY399 pKa = 10.79TPTAAYY405 pKa = 8.09TGTATISLGSADD417 pKa = 3.71QGNTGGGVLSDD428 pKa = 3.82SDD430 pKa = 4.38TINIQIGASRR440 pKa = 11.84YY441 pKa = 6.8QQGLDD446 pKa = 3.62GYY448 pKa = 9.17TGMEE452 pKa = 4.13DD453 pKa = 3.55TYY455 pKa = 10.96LDD457 pKa = 3.79SVTPDD462 pKa = 2.9SSYY465 pKa = 11.97GNASTVLVDD474 pKa = 4.39DD475 pKa = 5.53GSPVSQGLLRR485 pKa = 11.84FDD487 pKa = 5.25GIFGDD492 pKa = 4.65GVGQIPFGSTINSASLSFYY511 pKa = 10.96VINQDD516 pKa = 3.35AADD519 pKa = 3.99SVSLHH524 pKa = 5.8TMLTNWTEE532 pKa = 3.71NATWNTLGAGIQTNGVEE549 pKa = 4.35AVGTPVATFSAGASGWNTIGGLDD572 pKa = 3.69ASVQIWANGGANYY585 pKa = 9.89GWSLLTANPGADD597 pKa = 2.6SWTFASSEE605 pKa = 4.09YY606 pKa = 9.78STLNLRR612 pKa = 11.84PYY614 pKa = 10.85LVITYY619 pKa = 7.4TPPEE623 pKa = 4.19APVITSNGGGASGVVSVPEE642 pKa = 3.99NTTSVTTVTATDD654 pKa = 3.57ADD656 pKa = 3.93SAQTVLRR663 pKa = 11.84YY664 pKa = 10.2SIVGGADD671 pKa = 3.05STRR674 pKa = 11.84FSINEE679 pKa = 3.97TTGVLSFLTAPDD691 pKa = 4.02FEE693 pKa = 5.25NPLDD697 pKa = 3.56IGGNNIYY704 pKa = 10.43DD705 pKa = 3.43ITVQVSDD712 pKa = 4.52GLLSSTQVLAVSVGAVNDD730 pKa = 3.72NAPLITIPAAVWITEE745 pKa = 3.72NSTAVATITVSDD757 pKa = 4.05SDD759 pKa = 4.12RR760 pKa = 11.84PAQSFSYY767 pKa = 10.42SLAGGVDD774 pKa = 3.16AALFTINTSTGDD786 pKa = 3.76LRR788 pKa = 11.84FLSAPDD794 pKa = 3.76FEE796 pKa = 5.3SPLDD800 pKa = 3.84AGGDD804 pKa = 3.45NVYY807 pKa = 10.93DD808 pKa = 3.47VTVQVSDD815 pKa = 3.77SDD817 pKa = 4.28GSTSTQAIAVTVTSADD833 pKa = 3.33DD834 pKa = 3.58SAPVITSPSVVFVAEE849 pKa = 3.94NTTEE853 pKa = 3.89VMTITASDD861 pKa = 3.84ADD863 pKa = 3.89QPAQALSYY871 pKa = 10.47SLAGGADD878 pKa = 3.44VARR881 pKa = 11.84FTIDD885 pKa = 3.25PVSGILRR892 pKa = 11.84FVRR895 pKa = 11.84APGFGVPADD904 pKa = 3.71ANRR907 pKa = 11.84DD908 pKa = 3.36NVYY911 pKa = 10.96DD912 pKa = 3.96VIVRR916 pKa = 11.84VSDD919 pKa = 3.83GVLTGTQALSVSVEE933 pKa = 4.0NRR935 pKa = 11.84NEE937 pKa = 3.89MPQGRR942 pKa = 11.84DD943 pKa = 2.86GRR945 pKa = 11.84VQMLEE950 pKa = 4.38DD951 pKa = 3.7GTHH954 pKa = 6.06TFTLASFGFMDD965 pKa = 5.62ARR967 pKa = 11.84DD968 pKa = 3.83TPANNLAAVQIIAGPAKK985 pKa = 10.22GVLSLGGQVVANGQWITAGDD1005 pKa = 3.58IAAGLLSYY1013 pKa = 10.37TPAADD1018 pKa = 3.77GTGAAYY1024 pKa = 9.73AALTFRR1030 pKa = 11.84VRR1032 pKa = 11.84DD1033 pKa = 3.89DD1034 pKa = 3.63GGTAGGGSDD1043 pKa = 4.05TEE1045 pKa = 4.68TGSHH1049 pKa = 5.27TLVIGVTGVNDD1060 pKa = 3.76VPVGSADD1067 pKa = 3.42IYY1069 pKa = 11.23RR1070 pKa = 11.84LDD1072 pKa = 4.0EE1073 pKa = 4.89DD1074 pKa = 4.07SVLDD1078 pKa = 3.67VLAPGVLANDD1088 pKa = 4.27PDD1090 pKa = 4.51LDD1092 pKa = 4.25GDD1094 pKa = 3.95MLVAEE1099 pKa = 4.95LVSGPTNGTLVLAADD1114 pKa = 4.28GSFRR1118 pKa = 11.84YY1119 pKa = 7.94TPSANWSGTEE1129 pKa = 3.96SFSYY1133 pKa = 10.56RR1134 pKa = 11.84PFDD1137 pKa = 4.3GIVYY1141 pKa = 10.48GDD1143 pKa = 3.6TTQVILVVEE1152 pKa = 4.63AVNDD1156 pKa = 3.65APEE1159 pKa = 4.87IIHH1162 pKa = 7.12ASFKK1166 pKa = 10.58VPGGGVVVLNADD1178 pKa = 3.35MMIASDD1184 pKa = 4.11VDD1186 pKa = 3.9SLPTSIHH1193 pKa = 5.51FQVDD1197 pKa = 3.48QVDD1200 pKa = 3.54NGHH1203 pKa = 6.51FEE1205 pKa = 4.49LVSAPGQAIRR1215 pKa = 11.84QFSYY1219 pKa = 11.55AEE1221 pKa = 3.89LAAGLVV1227 pKa = 3.63
MM1 pKa = 7.58SGGGDD6 pKa = 3.3VTRR9 pKa = 11.84FTIDD13 pKa = 3.39PDD15 pKa = 3.49TGALSFLTAPDD26 pKa = 3.86FEE28 pKa = 4.7VPQDD32 pKa = 3.27EE33 pKa = 4.83GANNVYY39 pKa = 10.6DD40 pKa = 4.36VIVQVMDD47 pKa = 4.18GANSTAIQAITVTVTAVNDD66 pKa = 3.6NAPVLTSNGGGVMAAVTIAEE86 pKa = 4.41STTLVTTLTATDD98 pKa = 3.7SDD100 pKa = 4.53LPGDD104 pKa = 3.66TLYY107 pKa = 11.42YY108 pKa = 10.34SIVGGADD115 pKa = 2.98AAKK118 pKa = 10.0FAIDD122 pKa = 3.79TFSGALNFALAPDD135 pKa = 3.96YY136 pKa = 10.86DD137 pKa = 3.97LPGDD141 pKa = 3.65VGGDD145 pKa = 3.46NIYY148 pKa = 11.15DD149 pKa = 3.69VIVRR153 pKa = 11.84VDD155 pKa = 3.45DD156 pKa = 3.35GTYY159 pKa = 10.97AVNQSLAITVSNTNEE174 pKa = 3.89APAVTITSTSYY185 pKa = 9.72TATEE189 pKa = 3.81QSALVLGGTGLSISDD204 pKa = 4.08ADD206 pKa = 3.86AGSHH210 pKa = 5.08VVSARR215 pKa = 11.84LNVVSGTLSATAGSSGVTITGSGTASLVFDD245 pKa = 3.97GTLAQINDD253 pKa = 3.78FLAGKK258 pKa = 9.94LGASLTYY265 pKa = 10.55LIDD268 pKa = 4.02IDD270 pKa = 4.49TPPASDD276 pKa = 3.86TLTLAADD283 pKa = 4.26DD284 pKa = 4.9LGHH287 pKa = 6.6TGSGTAASGSKK298 pKa = 10.23SVLINITAVNDD309 pKa = 4.13GPANTTPGTQVTLVNTPLVFGVSHH333 pKa = 6.63GNALSVTDD341 pKa = 4.51SDD343 pKa = 4.42AAANPLQISLVATHH357 pKa = 6.43GTLTLGNTTGLTFVSGDD374 pKa = 3.37GAADD378 pKa = 3.25AGMTFTGTQASINAALEE395 pKa = 4.28GLSYY399 pKa = 10.79TPTAAYY405 pKa = 8.09TGTATISLGSADD417 pKa = 3.71QGNTGGGVLSDD428 pKa = 3.82SDD430 pKa = 4.38TINIQIGASRR440 pKa = 11.84YY441 pKa = 6.8QQGLDD446 pKa = 3.62GYY448 pKa = 9.17TGMEE452 pKa = 4.13DD453 pKa = 3.55TYY455 pKa = 10.96LDD457 pKa = 3.79SVTPDD462 pKa = 2.9SSYY465 pKa = 11.97GNASTVLVDD474 pKa = 4.39DD475 pKa = 5.53GSPVSQGLLRR485 pKa = 11.84FDD487 pKa = 5.25GIFGDD492 pKa = 4.65GVGQIPFGSTINSASLSFYY511 pKa = 10.96VINQDD516 pKa = 3.35AADD519 pKa = 3.99SVSLHH524 pKa = 5.8TMLTNWTEE532 pKa = 3.71NATWNTLGAGIQTNGVEE549 pKa = 4.35AVGTPVATFSAGASGWNTIGGLDD572 pKa = 3.69ASVQIWANGGANYY585 pKa = 9.89GWSLLTANPGADD597 pKa = 2.6SWTFASSEE605 pKa = 4.09YY606 pKa = 9.78STLNLRR612 pKa = 11.84PYY614 pKa = 10.85LVITYY619 pKa = 7.4TPPEE623 pKa = 4.19APVITSNGGGASGVVSVPEE642 pKa = 3.99NTTSVTTVTATDD654 pKa = 3.57ADD656 pKa = 3.93SAQTVLRR663 pKa = 11.84YY664 pKa = 10.2SIVGGADD671 pKa = 3.05STRR674 pKa = 11.84FSINEE679 pKa = 3.97TTGVLSFLTAPDD691 pKa = 4.02FEE693 pKa = 5.25NPLDD697 pKa = 3.56IGGNNIYY704 pKa = 10.43DD705 pKa = 3.43ITVQVSDD712 pKa = 4.52GLLSSTQVLAVSVGAVNDD730 pKa = 3.72NAPLITIPAAVWITEE745 pKa = 3.72NSTAVATITVSDD757 pKa = 4.05SDD759 pKa = 4.12RR760 pKa = 11.84PAQSFSYY767 pKa = 10.42SLAGGVDD774 pKa = 3.16AALFTINTSTGDD786 pKa = 3.76LRR788 pKa = 11.84FLSAPDD794 pKa = 3.76FEE796 pKa = 5.3SPLDD800 pKa = 3.84AGGDD804 pKa = 3.45NVYY807 pKa = 10.93DD808 pKa = 3.47VTVQVSDD815 pKa = 3.77SDD817 pKa = 4.28GSTSTQAIAVTVTSADD833 pKa = 3.33DD834 pKa = 3.58SAPVITSPSVVFVAEE849 pKa = 3.94NTTEE853 pKa = 3.89VMTITASDD861 pKa = 3.84ADD863 pKa = 3.89QPAQALSYY871 pKa = 10.47SLAGGADD878 pKa = 3.44VARR881 pKa = 11.84FTIDD885 pKa = 3.25PVSGILRR892 pKa = 11.84FVRR895 pKa = 11.84APGFGVPADD904 pKa = 3.71ANRR907 pKa = 11.84DD908 pKa = 3.36NVYY911 pKa = 10.96DD912 pKa = 3.96VIVRR916 pKa = 11.84VSDD919 pKa = 3.83GVLTGTQALSVSVEE933 pKa = 4.0NRR935 pKa = 11.84NEE937 pKa = 3.89MPQGRR942 pKa = 11.84DD943 pKa = 2.86GRR945 pKa = 11.84VQMLEE950 pKa = 4.38DD951 pKa = 3.7GTHH954 pKa = 6.06TFTLASFGFMDD965 pKa = 5.62ARR967 pKa = 11.84DD968 pKa = 3.83TPANNLAAVQIIAGPAKK985 pKa = 10.22GVLSLGGQVVANGQWITAGDD1005 pKa = 3.58IAAGLLSYY1013 pKa = 10.37TPAADD1018 pKa = 3.77GTGAAYY1024 pKa = 9.73AALTFRR1030 pKa = 11.84VRR1032 pKa = 11.84DD1033 pKa = 3.89DD1034 pKa = 3.63GGTAGGGSDD1043 pKa = 4.05TEE1045 pKa = 4.68TGSHH1049 pKa = 5.27TLVIGVTGVNDD1060 pKa = 3.76VPVGSADD1067 pKa = 3.42IYY1069 pKa = 11.23RR1070 pKa = 11.84LDD1072 pKa = 4.0EE1073 pKa = 4.89DD1074 pKa = 4.07SVLDD1078 pKa = 3.67VLAPGVLANDD1088 pKa = 4.27PDD1090 pKa = 4.51LDD1092 pKa = 4.25GDD1094 pKa = 3.95MLVAEE1099 pKa = 4.95LVSGPTNGTLVLAADD1114 pKa = 4.28GSFRR1118 pKa = 11.84YY1119 pKa = 7.94TPSANWSGTEE1129 pKa = 3.96SFSYY1133 pKa = 10.56RR1134 pKa = 11.84PFDD1137 pKa = 4.3GIVYY1141 pKa = 10.48GDD1143 pKa = 3.6TTQVILVVEE1152 pKa = 4.63AVNDD1156 pKa = 3.65APEE1159 pKa = 4.87IIHH1162 pKa = 7.12ASFKK1166 pKa = 10.58VPGGGVVVLNADD1178 pKa = 3.35MMIASDD1184 pKa = 4.11VDD1186 pKa = 3.9SLPTSIHH1193 pKa = 5.51FQVDD1197 pKa = 3.48QVDD1200 pKa = 3.54NGHH1203 pKa = 6.51FEE1205 pKa = 4.49LVSAPGQAIRR1215 pKa = 11.84QFSYY1219 pKa = 11.55AEE1221 pKa = 3.89LAAGLVV1227 pKa = 3.63
Molecular weight: 124.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6C2CYH3|A0A6C2CYH3_9RHOO Glutamate dehydrogenase OS=Zoogloea oleivorans OX=1552750 GN=ETQ85_10755 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.6RR14 pKa = 11.84THH16 pKa = 5.75GFLVRR21 pKa = 11.84MGTRR25 pKa = 11.84GGRR28 pKa = 11.84KK29 pKa = 8.93VIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.0GRR39 pKa = 11.84HH40 pKa = 4.92RR41 pKa = 11.84LAVV44 pKa = 3.37
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.6RR14 pKa = 11.84THH16 pKa = 5.75GFLVRR21 pKa = 11.84MGTRR25 pKa = 11.84GGRR28 pKa = 11.84KK29 pKa = 8.93VIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.0GRR39 pKa = 11.84HH40 pKa = 4.92RR41 pKa = 11.84LAVV44 pKa = 3.37
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1663900 |
24 |
4131 |
334.7 |
36.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.985 ± 0.042 | 0.981 ± 0.012 |
5.494 ± 0.024 | 5.76 ± 0.032 |
3.638 ± 0.021 | 8.155 ± 0.044 |
2.247 ± 0.017 | 4.828 ± 0.023 |
3.495 ± 0.035 | 10.951 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.267 ± 0.019 | 2.768 ± 0.023 |
5.079 ± 0.027 | 3.575 ± 0.021 |
6.996 ± 0.04 | 5.61 ± 0.03 |
5.165 ± 0.043 | 7.391 ± 0.031 |
1.358 ± 0.015 | 2.256 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
