
Fengkai orbivirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus; unclassified Orbivirus; Tibet orbivirus
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
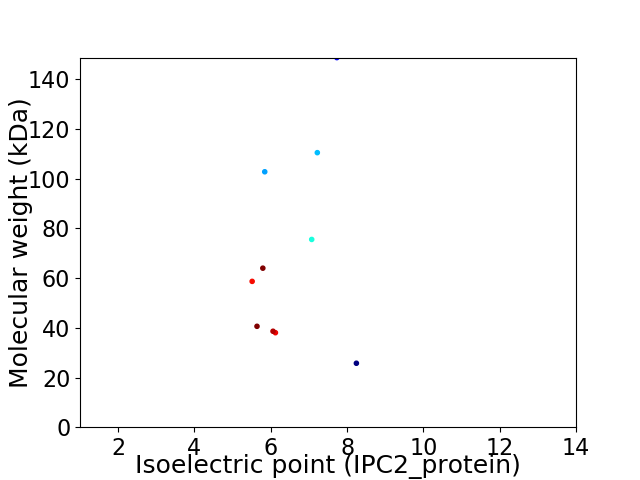
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0K1TPZ3|A0A0K1TPZ3_9REOV Non-structural protein NS1 OS=Fengkai orbivirus OX=1692107 GN=NS1 PE=4 SV=1
MM1 pKa = 6.78EE2 pKa = 5.71HH3 pKa = 6.36KK4 pKa = 10.33PKK6 pKa = 10.73RR7 pKa = 11.84FTKK10 pKa = 10.31NVSVLDD16 pKa = 4.06GSGTTICGIIAKK28 pKa = 9.39SAHH31 pKa = 6.37KK32 pKa = 9.26PYY34 pKa = 10.79CNISIGRR41 pKa = 11.84NVSIKK46 pKa = 10.69AVDD49 pKa = 3.74NPPPKK54 pKa = 10.44AYY56 pKa = 10.33VLQIRR61 pKa = 11.84QEE63 pKa = 3.84GGYY66 pKa = 9.99RR67 pKa = 11.84IQDD70 pKa = 3.65GQDD73 pKa = 3.45TISLMITASGVEE85 pKa = 3.99ATVEE89 pKa = 4.14RR90 pKa = 11.84WEE92 pKa = 4.0EE93 pKa = 3.74WQFEE97 pKa = 4.57VLSAMPMPMAIMMQHH112 pKa = 6.01NGEE115 pKa = 4.19MVDD118 pKa = 5.2AEE120 pKa = 4.11IKK122 pKa = 8.51YY123 pKa = 9.55AKK125 pKa = 10.36GMGVVAPYY133 pKa = 10.46VKK135 pKa = 10.37NDD137 pKa = 3.08EE138 pKa = 4.07VDD140 pKa = 3.37RR141 pKa = 11.84RR142 pKa = 11.84EE143 pKa = 4.12LPKK146 pKa = 10.9LPGLSEE152 pKa = 3.85SQYY155 pKa = 11.24DD156 pKa = 3.91VKK158 pKa = 10.15TLRR161 pKa = 11.84QKK163 pKa = 10.44IKK165 pKa = 9.53EE166 pKa = 3.94EE167 pKa = 3.97RR168 pKa = 11.84EE169 pKa = 3.86RR170 pKa = 11.84GVTEE174 pKa = 3.82IRR176 pKa = 11.84SKK178 pKa = 10.4IQPLEE183 pKa = 3.99QEE185 pKa = 4.52TKK187 pKa = 10.59RR188 pKa = 11.84MEE190 pKa = 4.27SVEE193 pKa = 4.1KK194 pKa = 10.93SFAEE198 pKa = 4.37MKK200 pKa = 10.54AIWAAEE206 pKa = 3.99SKK208 pKa = 11.29ADD210 pKa = 3.7VKK212 pKa = 11.05EE213 pKa = 4.17EE214 pKa = 4.24NVGGTYY220 pKa = 9.34GTDD223 pKa = 3.49FFKK226 pKa = 10.82PFPIEE231 pKa = 4.01STKK234 pKa = 10.2MPEE237 pKa = 4.23LKK239 pKa = 10.5SLEE242 pKa = 3.75QRR244 pKa = 11.84EE245 pKa = 4.12QQKK248 pKa = 10.43EE249 pKa = 4.21KK250 pKa = 10.77EE251 pKa = 4.33VAQAQSFSVDD261 pKa = 3.26VEE263 pKa = 4.48SGSEE267 pKa = 4.13EE268 pKa = 4.03EE269 pKa = 4.39VEE271 pKa = 4.09EE272 pKa = 4.21PNYY275 pKa = 9.03ITQEE279 pKa = 4.21YY280 pKa = 9.35VDD282 pKa = 4.1KK283 pKa = 10.92ASKK286 pKa = 9.56VVKK289 pKa = 10.5GGEE292 pKa = 3.99KK293 pKa = 10.06KK294 pKa = 10.15IAGLAYY300 pKa = 10.63VMPKK304 pKa = 10.46SIGDD308 pKa = 3.52FSKK311 pKa = 10.45IIAVKK316 pKa = 8.67KK317 pKa = 9.6HH318 pKa = 5.14KK319 pKa = 9.47WSNVPLFTVDD329 pKa = 3.59EE330 pKa = 4.72KK331 pKa = 11.29GLKK334 pKa = 10.16YY335 pKa = 10.04EE336 pKa = 4.07LQTIGEE342 pKa = 4.74CEE344 pKa = 4.18TVVYY348 pKa = 10.02AARR351 pKa = 11.84GLNLLVLPVGII362 pKa = 4.83
MM1 pKa = 6.78EE2 pKa = 5.71HH3 pKa = 6.36KK4 pKa = 10.33PKK6 pKa = 10.73RR7 pKa = 11.84FTKK10 pKa = 10.31NVSVLDD16 pKa = 4.06GSGTTICGIIAKK28 pKa = 9.39SAHH31 pKa = 6.37KK32 pKa = 9.26PYY34 pKa = 10.79CNISIGRR41 pKa = 11.84NVSIKK46 pKa = 10.69AVDD49 pKa = 3.74NPPPKK54 pKa = 10.44AYY56 pKa = 10.33VLQIRR61 pKa = 11.84QEE63 pKa = 3.84GGYY66 pKa = 9.99RR67 pKa = 11.84IQDD70 pKa = 3.65GQDD73 pKa = 3.45TISLMITASGVEE85 pKa = 3.99ATVEE89 pKa = 4.14RR90 pKa = 11.84WEE92 pKa = 4.0EE93 pKa = 3.74WQFEE97 pKa = 4.57VLSAMPMPMAIMMQHH112 pKa = 6.01NGEE115 pKa = 4.19MVDD118 pKa = 5.2AEE120 pKa = 4.11IKK122 pKa = 8.51YY123 pKa = 9.55AKK125 pKa = 10.36GMGVVAPYY133 pKa = 10.46VKK135 pKa = 10.37NDD137 pKa = 3.08EE138 pKa = 4.07VDD140 pKa = 3.37RR141 pKa = 11.84RR142 pKa = 11.84EE143 pKa = 4.12LPKK146 pKa = 10.9LPGLSEE152 pKa = 3.85SQYY155 pKa = 11.24DD156 pKa = 3.91VKK158 pKa = 10.15TLRR161 pKa = 11.84QKK163 pKa = 10.44IKK165 pKa = 9.53EE166 pKa = 3.94EE167 pKa = 3.97RR168 pKa = 11.84EE169 pKa = 3.86RR170 pKa = 11.84GVTEE174 pKa = 3.82IRR176 pKa = 11.84SKK178 pKa = 10.4IQPLEE183 pKa = 3.99QEE185 pKa = 4.52TKK187 pKa = 10.59RR188 pKa = 11.84MEE190 pKa = 4.27SVEE193 pKa = 4.1KK194 pKa = 10.93SFAEE198 pKa = 4.37MKK200 pKa = 10.54AIWAAEE206 pKa = 3.99SKK208 pKa = 11.29ADD210 pKa = 3.7VKK212 pKa = 11.05EE213 pKa = 4.17EE214 pKa = 4.24NVGGTYY220 pKa = 9.34GTDD223 pKa = 3.49FFKK226 pKa = 10.82PFPIEE231 pKa = 4.01STKK234 pKa = 10.2MPEE237 pKa = 4.23LKK239 pKa = 10.5SLEE242 pKa = 3.75QRR244 pKa = 11.84EE245 pKa = 4.12QQKK248 pKa = 10.43EE249 pKa = 4.21KK250 pKa = 10.77EE251 pKa = 4.33VAQAQSFSVDD261 pKa = 3.26VEE263 pKa = 4.48SGSEE267 pKa = 4.13EE268 pKa = 4.03EE269 pKa = 4.39VEE271 pKa = 4.09EE272 pKa = 4.21PNYY275 pKa = 9.03ITQEE279 pKa = 4.21YY280 pKa = 9.35VDD282 pKa = 4.1KK283 pKa = 10.92ASKK286 pKa = 9.56VVKK289 pKa = 10.5GGEE292 pKa = 3.99KK293 pKa = 10.06KK294 pKa = 10.15IAGLAYY300 pKa = 10.63VMPKK304 pKa = 10.46SIGDD308 pKa = 3.52FSKK311 pKa = 10.45IIAVKK316 pKa = 8.67KK317 pKa = 9.6HH318 pKa = 5.14KK319 pKa = 9.47WSNVPLFTVDD329 pKa = 3.59EE330 pKa = 4.72KK331 pKa = 11.29GLKK334 pKa = 10.16YY335 pKa = 10.04EE336 pKa = 4.07LQTIGEE342 pKa = 4.74CEE344 pKa = 4.18TVVYY348 pKa = 10.02AARR351 pKa = 11.84GLNLLVLPVGII362 pKa = 4.83
Molecular weight: 40.62 kDa
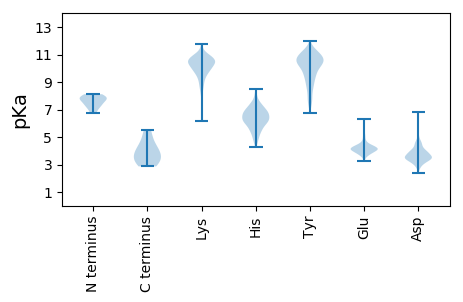
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1TPZ7|A0A0K1TPZ7_9REOV Non-structural protein NS3 OS=Fengkai orbivirus OX=1692107 GN=NS3 PE=3 SV=1
MM1 pKa = 7.72LSGLVRR7 pKa = 11.84RR8 pKa = 11.84FEE10 pKa = 4.15EE11 pKa = 4.26EE12 pKa = 4.12RR13 pKa = 11.84IEE15 pKa = 4.07MPQEE19 pKa = 3.92VKK21 pKa = 10.37PDD23 pKa = 3.52EE24 pKa = 4.57LSLVNYY30 pKa = 9.01YY31 pKa = 9.98PSEE34 pKa = 4.16SRR36 pKa = 11.84PPSYY40 pKa = 11.46APMAPTMTDD49 pKa = 2.4GMANVSLGILNQAMTNTTGATTASKK74 pKa = 10.72LEE76 pKa = 3.83KK77 pKa = 10.04AAYY80 pKa = 9.67GSYY83 pKa = 11.22AEE85 pKa = 4.89AFRR88 pKa = 11.84DD89 pKa = 3.9DD90 pKa = 3.47LRR92 pKa = 11.84LRR94 pKa = 11.84QVKK97 pKa = 9.85KK98 pKa = 10.12HH99 pKa = 6.07VNDD102 pKa = 3.67QIIPRR107 pKa = 11.84LSTEE111 pKa = 3.89LSHH114 pKa = 7.45LKK116 pKa = 9.9RR117 pKa = 11.84KK118 pKa = 9.95KK119 pKa = 10.35IIVGTIMLVAAIVALVTSAGTLTNDD144 pKa = 4.43LSISMKK150 pKa = 10.75AGDD153 pKa = 3.98VNITIPAWFKK163 pKa = 10.84GFSAVFGFANLGSTMIMMLCARR185 pKa = 11.84TEE187 pKa = 3.87RR188 pKa = 11.84MLASQIEE195 pKa = 4.58MIQKK199 pKa = 9.96EE200 pKa = 4.33LMKK203 pKa = 10.53KK204 pKa = 10.1RR205 pKa = 11.84SYY207 pKa = 11.28NEE209 pKa = 3.65AVRR212 pKa = 11.84MSTAEE217 pKa = 3.85LSGMSLEE224 pKa = 4.94GIIPNTGQLTT234 pKa = 3.68
MM1 pKa = 7.72LSGLVRR7 pKa = 11.84RR8 pKa = 11.84FEE10 pKa = 4.15EE11 pKa = 4.26EE12 pKa = 4.12RR13 pKa = 11.84IEE15 pKa = 4.07MPQEE19 pKa = 3.92VKK21 pKa = 10.37PDD23 pKa = 3.52EE24 pKa = 4.57LSLVNYY30 pKa = 9.01YY31 pKa = 9.98PSEE34 pKa = 4.16SRR36 pKa = 11.84PPSYY40 pKa = 11.46APMAPTMTDD49 pKa = 2.4GMANVSLGILNQAMTNTTGATTASKK74 pKa = 10.72LEE76 pKa = 3.83KK77 pKa = 10.04AAYY80 pKa = 9.67GSYY83 pKa = 11.22AEE85 pKa = 4.89AFRR88 pKa = 11.84DD89 pKa = 3.9DD90 pKa = 3.47LRR92 pKa = 11.84LRR94 pKa = 11.84QVKK97 pKa = 9.85KK98 pKa = 10.12HH99 pKa = 6.07VNDD102 pKa = 3.67QIIPRR107 pKa = 11.84LSTEE111 pKa = 3.89LSHH114 pKa = 7.45LKK116 pKa = 9.9RR117 pKa = 11.84KK118 pKa = 9.95KK119 pKa = 10.35IIVGTIMLVAAIVALVTSAGTLTNDD144 pKa = 4.43LSISMKK150 pKa = 10.75AGDD153 pKa = 3.98VNITIPAWFKK163 pKa = 10.84GFSAVFGFANLGSTMIMMLCARR185 pKa = 11.84TEE187 pKa = 3.87RR188 pKa = 11.84MLASQIEE195 pKa = 4.58MIQKK199 pKa = 9.96EE200 pKa = 4.33LMKK203 pKa = 10.53KK204 pKa = 10.1RR205 pKa = 11.84SYY207 pKa = 11.28NEE209 pKa = 3.65AVRR212 pKa = 11.84MSTAEE217 pKa = 3.85LSGMSLEE224 pKa = 4.94GIIPNTGQLTT234 pKa = 3.68
Molecular weight: 25.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6167 |
234 |
1304 |
616.7 |
70.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.297 ± 0.529 | 1.005 ± 0.298 |
5.919 ± 0.333 | 7.508 ± 0.752 |
3.616 ± 0.339 | 5.951 ± 0.363 |
2.416 ± 0.222 | 6.713 ± 0.445 |
6.535 ± 0.899 | 8.659 ± 0.455 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.892 ± 0.242 | 3.357 ± 0.206 |
3.859 ± 0.361 | 3.746 ± 0.299 |
6.502 ± 0.418 | 5.643 ± 0.419 |
5.254 ± 0.385 | 6.94 ± 0.349 |
1.297 ± 0.234 | 3.892 ± 0.383 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
