
Polymorphum gilvum (strain LMG 25793 / CGMCC 1.9160 / SL003B-26A1)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Polymorphum; Polymorphum gilvum
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
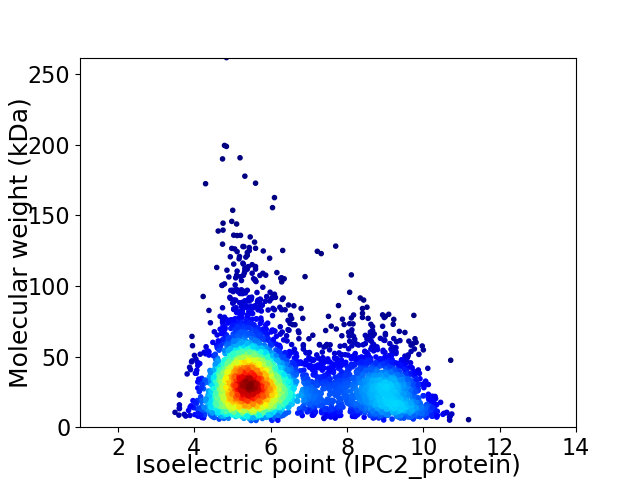
Virtual 2D-PAGE plot for 4354 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F2J431|F2J431_POLGS Uncharacterized protein OS=Polymorphum gilvum (strain LMG 25793 / CGMCC 1.9160 / SL003B-26A1) OX=991905 GN=SL003B_1529 PE=4 SV=1
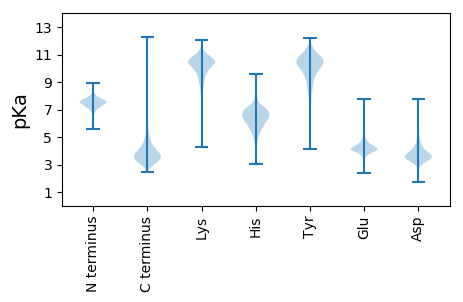
MM1 pKa = 6.54TQADD5 pKa = 4.1AVIDD9 pKa = 4.23GQTTFATTEE18 pKa = 4.15YY19 pKa = 8.68YY20 pKa = 9.98TRR22 pKa = 11.84AGDD25 pKa = 3.99FEE27 pKa = 5.68LNEE30 pKa = 4.65DD31 pKa = 3.94GYY33 pKa = 10.13MVNSAGYY40 pKa = 8.9YY41 pKa = 10.79LMGFPLDD48 pKa = 3.62ARR50 pKa = 11.84TGNAVGDD57 pKa = 4.05NPEE60 pKa = 4.26VIQIGSQPLPAQATSEE76 pKa = 3.99IKK78 pKa = 10.68YY79 pKa = 9.51QANLPSYY86 pKa = 9.1PQTNNADD93 pKa = 3.5TSVPGSQYY101 pKa = 11.09LDD103 pKa = 3.13TSGAITTTSDD113 pKa = 2.87IAVADD118 pKa = 3.44IDD120 pKa = 5.08DD121 pKa = 4.64FLNSSISGQSITVYY135 pKa = 10.23DD136 pKa = 3.95QNGTPLNVEE145 pKa = 4.12VRR147 pKa = 11.84WAQTAANTWNAYY159 pKa = 9.19YY160 pKa = 10.68SSGGDD165 pKa = 3.85AVTGWYY171 pKa = 10.46LIGEE175 pKa = 4.54VTFDD179 pKa = 3.25AAGVMDD185 pKa = 4.89TLTPAGTNTVGAGGASFVIDD205 pKa = 4.33DD206 pKa = 4.07LTVAGVNAGDD216 pKa = 3.92IEE218 pKa = 4.54FSFKK222 pKa = 10.47KK223 pKa = 10.77GSLTQYY229 pKa = 11.15ADD231 pKa = 3.56PKK233 pKa = 10.41GLASSVSLNQDD244 pKa = 2.8GYY246 pKa = 11.71AAGQVVGLSIDD257 pKa = 3.38GAGRR261 pKa = 11.84IIASYY266 pKa = 11.02SNQQTRR272 pKa = 11.84SLFEE276 pKa = 4.12IPLATFQADD285 pKa = 3.38QNLRR289 pKa = 11.84RR290 pKa = 11.84VDD292 pKa = 3.78GSAFAATPSSGKK304 pKa = 9.9AAYY307 pKa = 10.01GNGGTIVANALEE319 pKa = 4.44SSNADD324 pKa = 2.88IAAEE328 pKa = 3.99FSKK331 pKa = 11.23LIITQQAYY339 pKa = 9.4SANSKK344 pKa = 10.15VVTSADD350 pKa = 3.24QMLNDD355 pKa = 4.43ALNMVRR361 pKa = 5.43
MM1 pKa = 6.54TQADD5 pKa = 4.1AVIDD9 pKa = 4.23GQTTFATTEE18 pKa = 4.15YY19 pKa = 8.68YY20 pKa = 9.98TRR22 pKa = 11.84AGDD25 pKa = 3.99FEE27 pKa = 5.68LNEE30 pKa = 4.65DD31 pKa = 3.94GYY33 pKa = 10.13MVNSAGYY40 pKa = 8.9YY41 pKa = 10.79LMGFPLDD48 pKa = 3.62ARR50 pKa = 11.84TGNAVGDD57 pKa = 4.05NPEE60 pKa = 4.26VIQIGSQPLPAQATSEE76 pKa = 3.99IKK78 pKa = 10.68YY79 pKa = 9.51QANLPSYY86 pKa = 9.1PQTNNADD93 pKa = 3.5TSVPGSQYY101 pKa = 11.09LDD103 pKa = 3.13TSGAITTTSDD113 pKa = 2.87IAVADD118 pKa = 3.44IDD120 pKa = 5.08DD121 pKa = 4.64FLNSSISGQSITVYY135 pKa = 10.23DD136 pKa = 3.95QNGTPLNVEE145 pKa = 4.12VRR147 pKa = 11.84WAQTAANTWNAYY159 pKa = 9.19YY160 pKa = 10.68SSGGDD165 pKa = 3.85AVTGWYY171 pKa = 10.46LIGEE175 pKa = 4.54VTFDD179 pKa = 3.25AAGVMDD185 pKa = 4.89TLTPAGTNTVGAGGASFVIDD205 pKa = 4.33DD206 pKa = 4.07LTVAGVNAGDD216 pKa = 3.92IEE218 pKa = 4.54FSFKK222 pKa = 10.47KK223 pKa = 10.77GSLTQYY229 pKa = 11.15ADD231 pKa = 3.56PKK233 pKa = 10.41GLASSVSLNQDD244 pKa = 2.8GYY246 pKa = 11.71AAGQVVGLSIDD257 pKa = 3.38GAGRR261 pKa = 11.84IIASYY266 pKa = 11.02SNQQTRR272 pKa = 11.84SLFEE276 pKa = 4.12IPLATFQADD285 pKa = 3.38QNLRR289 pKa = 11.84RR290 pKa = 11.84VDD292 pKa = 3.78GSAFAATPSSGKK304 pKa = 9.9AAYY307 pKa = 10.01GNGGTIVANALEE319 pKa = 4.44SSNADD324 pKa = 2.88IAAEE328 pKa = 3.99FSKK331 pKa = 11.23LIITQQAYY339 pKa = 9.4SANSKK344 pKa = 10.15VVTSADD350 pKa = 3.24QMLNDD355 pKa = 4.43ALNMVRR361 pKa = 5.43
Molecular weight: 37.71 kDa
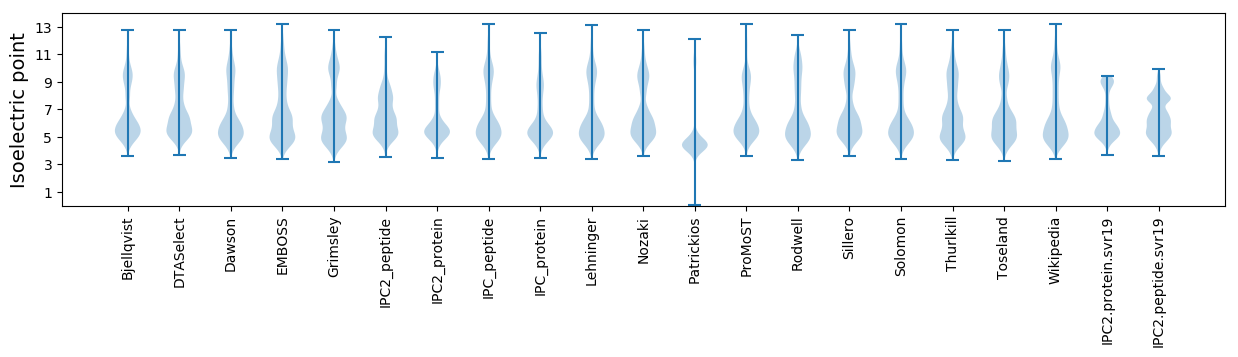
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F2J217|F2J217_POLGS von Willebrand factor type A:Cobalt chelatase pCobT subunit OS=Polymorphum gilvum (strain LMG 25793 / CGMCC 1.9160 / SL003B-26A1) OX=991905 GN=SL003B_0341 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.48GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.29NGRR28 pKa = 11.84QVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.99GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.48GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.29NGRR28 pKa = 11.84QVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.99GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1377863 |
39 |
2444 |
316.5 |
34.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.342 ± 0.055 | 0.843 ± 0.011 |
5.997 ± 0.029 | 5.77 ± 0.031 |
3.647 ± 0.024 | 8.705 ± 0.042 |
1.979 ± 0.017 | 4.849 ± 0.027 |
2.836 ± 0.026 | 10.374 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.369 ± 0.018 | 2.221 ± 0.018 |
5.173 ± 0.028 | 2.829 ± 0.019 |
7.747 ± 0.045 | 4.966 ± 0.022 |
5.261 ± 0.025 | 7.68 ± 0.032 |
1.281 ± 0.014 | 2.131 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
