
Avon-Heathcote Estuary associated circular virus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
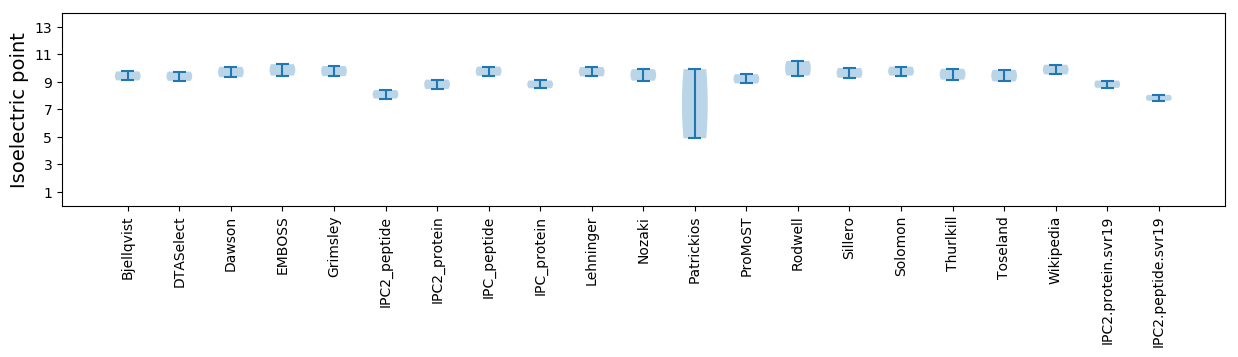
Average proteome isoelectric point is 8.83
Get precalculated fractions of proteins
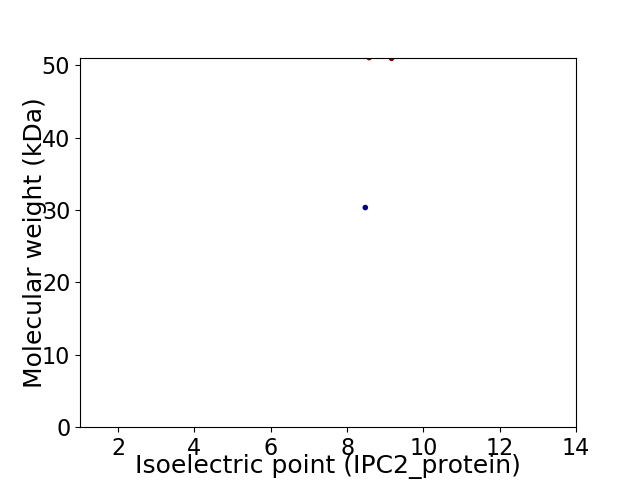
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IM76|A0A0C5IM76_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 3 OX=1618254 PE=4 SV=1
MM1 pKa = 6.69NQARR5 pKa = 11.84FWILTIRR12 pKa = 11.84HH13 pKa = 6.39ADD15 pKa = 3.71FLPYY19 pKa = 10.48LPPTVDD25 pKa = 3.3YY26 pKa = 11.34LRR28 pKa = 11.84GQLEE32 pKa = 4.07RR33 pKa = 11.84GDD35 pKa = 4.06GGFLHH40 pKa = 6.48WQLVVHH46 pKa = 6.65FARR49 pKa = 11.84KK50 pKa = 9.04VRR52 pKa = 11.84LGGVKK57 pKa = 10.46GIFGDD62 pKa = 3.95STHH65 pKa = 7.16AEE67 pKa = 4.19PTRR70 pKa = 11.84SDD72 pKa = 3.15AARR75 pKa = 11.84EE76 pKa = 4.1YY77 pKa = 10.4VWKK80 pKa = 10.24EE81 pKa = 3.51DD82 pKa = 3.4SAVPNTRR89 pKa = 11.84FEE91 pKa = 4.36LGKK94 pKa = 9.74IPVRR98 pKa = 11.84RR99 pKa = 11.84GVSHH103 pKa = 7.2DD104 pKa = 2.91WDD106 pKa = 3.68AVRR109 pKa = 11.84EE110 pKa = 4.19SAKK113 pKa = 10.31RR114 pKa = 11.84GRR116 pKa = 11.84LDD118 pKa = 5.85DD119 pKa = 3.99IPADD123 pKa = 3.87IYY125 pKa = 11.23CRR127 pKa = 11.84LYY129 pKa = 11.48GNFKK133 pKa = 10.52RR134 pKa = 11.84IAVDD138 pKa = 3.03HH139 pKa = 5.89MVPLGIEE146 pKa = 4.06RR147 pKa = 11.84EE148 pKa = 4.34VNVYY152 pKa = 9.44WGVTGSGKK160 pKa = 8.79SRR162 pKa = 11.84RR163 pKa = 11.84AWNEE167 pKa = 3.19AGLDD171 pKa = 3.85AFPKK175 pKa = 10.49DD176 pKa = 3.69PRR178 pKa = 11.84TKK180 pKa = 10.17FWDD183 pKa = 4.42GYY185 pKa = 9.57RR186 pKa = 11.84GHH188 pKa = 6.83EE189 pKa = 3.85NVVIDD194 pKa = 4.25EE195 pKa = 4.26FRR197 pKa = 11.84GGIDD201 pKa = 3.25VAHH204 pKa = 7.59LLRR207 pKa = 11.84WFDD210 pKa = 3.74RR211 pKa = 11.84YY212 pKa = 10.05PVVVEE217 pKa = 4.03VKK219 pKa = 10.13GSSVVLSAKK228 pKa = 10.15HH229 pKa = 5.47IWITSNLDD237 pKa = 3.12PRR239 pKa = 11.84EE240 pKa = 4.04WYY242 pKa = 10.5ADD244 pKa = 3.54LDD246 pKa = 4.21AEE248 pKa = 4.38TLAALLRR255 pKa = 11.84RR256 pKa = 11.84LKK258 pKa = 9.24ITQFAII264 pKa = 3.58
MM1 pKa = 6.69NQARR5 pKa = 11.84FWILTIRR12 pKa = 11.84HH13 pKa = 6.39ADD15 pKa = 3.71FLPYY19 pKa = 10.48LPPTVDD25 pKa = 3.3YY26 pKa = 11.34LRR28 pKa = 11.84GQLEE32 pKa = 4.07RR33 pKa = 11.84GDD35 pKa = 4.06GGFLHH40 pKa = 6.48WQLVVHH46 pKa = 6.65FARR49 pKa = 11.84KK50 pKa = 9.04VRR52 pKa = 11.84LGGVKK57 pKa = 10.46GIFGDD62 pKa = 3.95STHH65 pKa = 7.16AEE67 pKa = 4.19PTRR70 pKa = 11.84SDD72 pKa = 3.15AARR75 pKa = 11.84EE76 pKa = 4.1YY77 pKa = 10.4VWKK80 pKa = 10.24EE81 pKa = 3.51DD82 pKa = 3.4SAVPNTRR89 pKa = 11.84FEE91 pKa = 4.36LGKK94 pKa = 9.74IPVRR98 pKa = 11.84RR99 pKa = 11.84GVSHH103 pKa = 7.2DD104 pKa = 2.91WDD106 pKa = 3.68AVRR109 pKa = 11.84EE110 pKa = 4.19SAKK113 pKa = 10.31RR114 pKa = 11.84GRR116 pKa = 11.84LDD118 pKa = 5.85DD119 pKa = 3.99IPADD123 pKa = 3.87IYY125 pKa = 11.23CRR127 pKa = 11.84LYY129 pKa = 11.48GNFKK133 pKa = 10.52RR134 pKa = 11.84IAVDD138 pKa = 3.03HH139 pKa = 5.89MVPLGIEE146 pKa = 4.06RR147 pKa = 11.84EE148 pKa = 4.34VNVYY152 pKa = 9.44WGVTGSGKK160 pKa = 8.79SRR162 pKa = 11.84RR163 pKa = 11.84AWNEE167 pKa = 3.19AGLDD171 pKa = 3.85AFPKK175 pKa = 10.49DD176 pKa = 3.69PRR178 pKa = 11.84TKK180 pKa = 10.17FWDD183 pKa = 4.42GYY185 pKa = 9.57RR186 pKa = 11.84GHH188 pKa = 6.83EE189 pKa = 3.85NVVIDD194 pKa = 4.25EE195 pKa = 4.26FRR197 pKa = 11.84GGIDD201 pKa = 3.25VAHH204 pKa = 7.59LLRR207 pKa = 11.84WFDD210 pKa = 3.74RR211 pKa = 11.84YY212 pKa = 10.05PVVVEE217 pKa = 4.03VKK219 pKa = 10.13GSSVVLSAKK228 pKa = 10.15HH229 pKa = 5.47IWITSNLDD237 pKa = 3.12PRR239 pKa = 11.84EE240 pKa = 4.04WYY242 pKa = 10.5ADD244 pKa = 3.54LDD246 pKa = 4.21AEE248 pKa = 4.38TLAALLRR255 pKa = 11.84RR256 pKa = 11.84LKK258 pKa = 9.24ITQFAII264 pKa = 3.58
Molecular weight: 30.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IM76|A0A0C5IM76_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 3 OX=1618254 PE=4 SV=1
MM1 pKa = 7.28AGKK4 pKa = 10.09RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84SFSGNVRR14 pKa = 11.84SVTRR18 pKa = 11.84VIPGPYY24 pKa = 8.91TPRR27 pKa = 11.84TTVGRR32 pKa = 11.84RR33 pKa = 11.84PATGFISGAASVARR47 pKa = 11.84QLAPQFARR55 pKa = 11.84IAAEE59 pKa = 3.95AATQTAINYY68 pKa = 6.36VTGGTQTKK76 pKa = 9.23TKK78 pKa = 9.4TKK80 pKa = 10.32SKK82 pKa = 9.84NKK84 pKa = 10.02SSTGMNVRR92 pKa = 11.84TSMSGGFFGNKK103 pKa = 5.8QKK105 pKa = 11.35KK106 pKa = 10.29NIFQKK111 pKa = 8.61YY112 pKa = 3.68TTRR115 pKa = 11.84GIVTTKK121 pKa = 8.62EE122 pKa = 3.74TANILIGGPTDD133 pKa = 3.64PDD135 pKa = 3.55LPVFFGLATHH145 pKa = 7.27ANFLLIGRR153 pKa = 11.84LFFRR157 pKa = 11.84TVVKK161 pKa = 10.69DD162 pKa = 3.49VMRR165 pKa = 11.84KK166 pKa = 9.51SGMNVSSMSDD176 pKa = 3.34TPQGGIFQLQWQIRR190 pKa = 11.84DD191 pKa = 3.17VDD193 pKa = 4.04DD194 pKa = 4.63AQSSGASVNTAGKK207 pKa = 7.38TLEE210 pKa = 4.33VIADD214 pKa = 3.95EE215 pKa = 5.54LYY217 pKa = 11.09ADD219 pKa = 3.72YY220 pKa = 11.39LSFFRR225 pKa = 11.84STLNAMEE232 pKa = 4.65LAYY235 pKa = 9.83ISSAQFWTPSFGTLLAQIDD254 pKa = 4.1LKK256 pKa = 10.69DD257 pKa = 3.6ARR259 pKa = 11.84IHH261 pKa = 6.03ILCKK265 pKa = 10.12SDD267 pKa = 5.09LKK269 pKa = 11.05AQNRR273 pKa = 11.84TLANGADD280 pKa = 4.02DD281 pKa = 5.81DD282 pKa = 4.98NNTTEE287 pKa = 4.7NIEE290 pKa = 3.94NQPLYY295 pKa = 10.5GKK297 pKa = 10.04QYY299 pKa = 8.75FGKK302 pKa = 10.6GVGLVTRR309 pKa = 11.84IGLNTTNASEE319 pKa = 4.39GKK321 pKa = 10.22SLICSQQTGLLSFAVKK337 pKa = 9.0KK338 pKa = 9.39TGTPGVNQEE347 pKa = 3.31PWLRR351 pKa = 11.84EE352 pKa = 3.84VPLPTLFCGKK362 pKa = 9.62IAYY365 pKa = 8.85GKK367 pKa = 10.56VSIEE371 pKa = 3.76PGQVKK376 pKa = 9.51TSSLTHH382 pKa = 6.34KK383 pKa = 10.83ASIKK387 pKa = 10.29QQDD390 pKa = 3.14FWNYY394 pKa = 9.5LRR396 pKa = 11.84EE397 pKa = 4.12NRR399 pKa = 11.84STAVKK404 pKa = 10.14GGRR407 pKa = 11.84PEE409 pKa = 3.42KK410 pKa = 10.15MGIFRR415 pKa = 11.84IFCVEE420 pKa = 4.71KK421 pKa = 10.07MICCTAEE428 pKa = 3.65DD429 pKa = 4.22RR430 pKa = 11.84APQLGIEE437 pKa = 4.2LNQRR441 pKa = 11.84IGLYY445 pKa = 9.29VAYY448 pKa = 9.26PRR450 pKa = 11.84VKK452 pKa = 9.57NTMEE456 pKa = 4.04MYY458 pKa = 10.46DD459 pKa = 3.38GHH461 pKa = 7.03EE462 pKa = 4.24YY463 pKa = 9.59TT464 pKa = 5.4
MM1 pKa = 7.28AGKK4 pKa = 10.09RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84SFSGNVRR14 pKa = 11.84SVTRR18 pKa = 11.84VIPGPYY24 pKa = 8.91TPRR27 pKa = 11.84TTVGRR32 pKa = 11.84RR33 pKa = 11.84PATGFISGAASVARR47 pKa = 11.84QLAPQFARR55 pKa = 11.84IAAEE59 pKa = 3.95AATQTAINYY68 pKa = 6.36VTGGTQTKK76 pKa = 9.23TKK78 pKa = 9.4TKK80 pKa = 10.32SKK82 pKa = 9.84NKK84 pKa = 10.02SSTGMNVRR92 pKa = 11.84TSMSGGFFGNKK103 pKa = 5.8QKK105 pKa = 11.35KK106 pKa = 10.29NIFQKK111 pKa = 8.61YY112 pKa = 3.68TTRR115 pKa = 11.84GIVTTKK121 pKa = 8.62EE122 pKa = 3.74TANILIGGPTDD133 pKa = 3.64PDD135 pKa = 3.55LPVFFGLATHH145 pKa = 7.27ANFLLIGRR153 pKa = 11.84LFFRR157 pKa = 11.84TVVKK161 pKa = 10.69DD162 pKa = 3.49VMRR165 pKa = 11.84KK166 pKa = 9.51SGMNVSSMSDD176 pKa = 3.34TPQGGIFQLQWQIRR190 pKa = 11.84DD191 pKa = 3.17VDD193 pKa = 4.04DD194 pKa = 4.63AQSSGASVNTAGKK207 pKa = 7.38TLEE210 pKa = 4.33VIADD214 pKa = 3.95EE215 pKa = 5.54LYY217 pKa = 11.09ADD219 pKa = 3.72YY220 pKa = 11.39LSFFRR225 pKa = 11.84STLNAMEE232 pKa = 4.65LAYY235 pKa = 9.83ISSAQFWTPSFGTLLAQIDD254 pKa = 4.1LKK256 pKa = 10.69DD257 pKa = 3.6ARR259 pKa = 11.84IHH261 pKa = 6.03ILCKK265 pKa = 10.12SDD267 pKa = 5.09LKK269 pKa = 11.05AQNRR273 pKa = 11.84TLANGADD280 pKa = 4.02DD281 pKa = 5.81DD282 pKa = 4.98NNTTEE287 pKa = 4.7NIEE290 pKa = 3.94NQPLYY295 pKa = 10.5GKK297 pKa = 10.04QYY299 pKa = 8.75FGKK302 pKa = 10.6GVGLVTRR309 pKa = 11.84IGLNTTNASEE319 pKa = 4.39GKK321 pKa = 10.22SLICSQQTGLLSFAVKK337 pKa = 9.0KK338 pKa = 9.39TGTPGVNQEE347 pKa = 3.31PWLRR351 pKa = 11.84EE352 pKa = 3.84VPLPTLFCGKK362 pKa = 9.62IAYY365 pKa = 8.85GKK367 pKa = 10.56VSIEE371 pKa = 3.76PGQVKK376 pKa = 9.51TSSLTHH382 pKa = 6.34KK383 pKa = 10.83ASIKK387 pKa = 10.29QQDD390 pKa = 3.14FWNYY394 pKa = 9.5LRR396 pKa = 11.84EE397 pKa = 4.12NRR399 pKa = 11.84STAVKK404 pKa = 10.14GGRR407 pKa = 11.84PEE409 pKa = 3.42KK410 pKa = 10.15MGIFRR415 pKa = 11.84IFCVEE420 pKa = 4.71KK421 pKa = 10.07MICCTAEE428 pKa = 3.65DD429 pKa = 4.22RR430 pKa = 11.84APQLGIEE437 pKa = 4.2LNQRR441 pKa = 11.84IGLYY445 pKa = 9.29VAYY448 pKa = 9.26PRR450 pKa = 11.84VKK452 pKa = 9.57NTMEE456 pKa = 4.04MYY458 pKa = 10.46DD459 pKa = 3.38GHH461 pKa = 7.03EE462 pKa = 4.24YY463 pKa = 9.59TT464 pKa = 5.4
Molecular weight: 50.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
728 |
264 |
464 |
364.0 |
40.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.83 ± 0.068 | 0.962 ± 0.315 |
5.357 ± 1.405 | 4.396 ± 0.491 |
4.67 ± 0.068 | 8.791 ± 0.248 |
1.786 ± 0.878 | 5.632 ± 0.178 |
5.907 ± 0.736 | 7.555 ± 0.421 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.786 ± 0.556 | 4.258 ± 0.869 |
4.258 ± 0.155 | 3.846 ± 1.261 |
7.555 ± 1.446 | 5.907 ± 0.941 |
7.418 ± 1.964 | 7.005 ± 1.128 |
1.923 ± 1.009 | 3.159 ± 0.135 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
