
Torque teno mini virus 8
Taxonomy: Viruses; Anelloviridae; Betatorquevirus
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
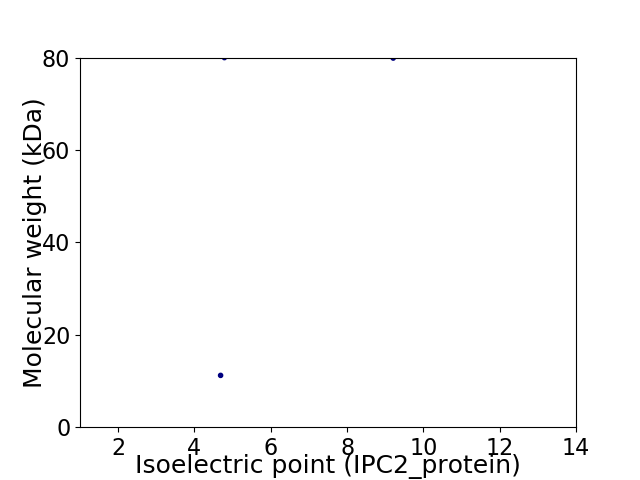
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
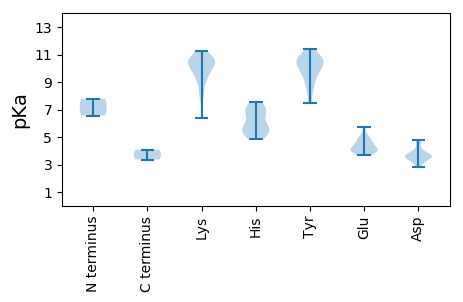
Protein with the lowest isoelectric point:
>tr|Q91C94|Q91C94_9VIRU ORF2 OS=Torque teno mini virus 8 OX=687376 PE=4 SV=1
MM1 pKa = 6.56STRR4 pKa = 11.84YY5 pKa = 9.75IPTKK9 pKa = 10.29DD10 pKa = 2.9SHH12 pKa = 5.69RR13 pKa = 11.84QKK15 pKa = 10.86NLKK18 pKa = 8.19WLNFVTHH25 pKa = 6.29AHH27 pKa = 7.53DD28 pKa = 4.22IFCDD32 pKa = 3.76CDD34 pKa = 3.82SPLQHH39 pKa = 5.8TVANILIQEE48 pKa = 4.2PNIKK52 pKa = 10.44FNTQEE57 pKa = 3.75KK58 pKa = 10.72DD59 pKa = 3.46LLKK62 pKa = 10.67KK63 pKa = 10.76CLGEE67 pKa = 4.12TTTTEE72 pKa = 4.1GAGDD76 pKa = 3.76PEE78 pKa = 4.62GFGDD82 pKa = 4.61GDD84 pKa = 4.02LEE86 pKa = 4.58RR87 pKa = 11.84LFTEE91 pKa = 5.07DD92 pKa = 4.61FGEE95 pKa = 4.33EE96 pKa = 3.96DD97 pKa = 3.19TGG99 pKa = 4.08
MM1 pKa = 6.56STRR4 pKa = 11.84YY5 pKa = 9.75IPTKK9 pKa = 10.29DD10 pKa = 2.9SHH12 pKa = 5.69RR13 pKa = 11.84QKK15 pKa = 10.86NLKK18 pKa = 8.19WLNFVTHH25 pKa = 6.29AHH27 pKa = 7.53DD28 pKa = 4.22IFCDD32 pKa = 3.76CDD34 pKa = 3.82SPLQHH39 pKa = 5.8TVANILIQEE48 pKa = 4.2PNIKK52 pKa = 10.44FNTQEE57 pKa = 3.75KK58 pKa = 10.72DD59 pKa = 3.46LLKK62 pKa = 10.67KK63 pKa = 10.76CLGEE67 pKa = 4.12TTTTEE72 pKa = 4.1GAGDD76 pKa = 3.76PEE78 pKa = 4.62GFGDD82 pKa = 4.61GDD84 pKa = 4.02LEE86 pKa = 4.58RR87 pKa = 11.84LFTEE91 pKa = 5.07DD92 pKa = 4.61FGEE95 pKa = 4.33EE96 pKa = 3.96DD97 pKa = 3.19TGG99 pKa = 4.08
Molecular weight: 11.22 kDa
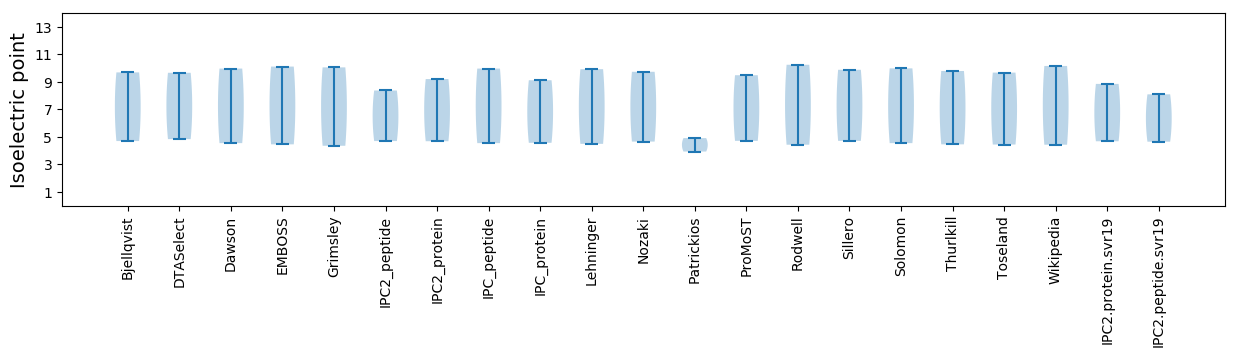
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q91C94|Q91C94_9VIRU ORF2 OS=Torque teno mini virus 8 OX=687376 PE=4 SV=1
MM1 pKa = 7.79PWRR4 pKa = 11.84NYY6 pKa = 8.37YY7 pKa = 8.77YY8 pKa = 10.24RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84PRR15 pKa = 11.84RR16 pKa = 11.84FWRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84FRR23 pKa = 11.84TPIYY27 pKa = 9.42RR28 pKa = 11.84RR29 pKa = 11.84FWRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84YY35 pKa = 9.38RR36 pKa = 11.84VRR38 pKa = 11.84KK39 pKa = 8.82RR40 pKa = 11.84HH41 pKa = 5.88FKK43 pKa = 10.65RR44 pKa = 11.84KK45 pKa = 9.28LSKK48 pKa = 10.04LTIQEE53 pKa = 4.02WQPPSIRR60 pKa = 11.84KK61 pKa = 8.27LTIKK65 pKa = 10.42GYY67 pKa = 10.22YY68 pKa = 9.53PLFQSTRR75 pKa = 11.84DD76 pKa = 3.43RR77 pKa = 11.84LSNNMITYY85 pKa = 10.1LEE87 pKa = 4.88SIAPHH92 pKa = 5.51YY93 pKa = 10.59VPGGGGFSISCFSLNTLYY111 pKa = 10.67KK112 pKa = 10.53EE113 pKa = 4.07HH114 pKa = 7.13LKK116 pKa = 10.5LHH118 pKa = 5.54NWWTKK123 pKa = 10.15TNDD126 pKa = 3.19NYY128 pKa = 11.09PLIRR132 pKa = 11.84YY133 pKa = 7.46NGCTLKK139 pKa = 10.73LFKK142 pKa = 10.43SEE144 pKa = 3.83KK145 pKa = 9.1VDD147 pKa = 3.57YY148 pKa = 10.31IFYY151 pKa = 9.01YY152 pKa = 9.07QRR154 pKa = 11.84HH155 pKa = 5.38YY156 pKa = 10.69PMKK159 pKa = 10.46ASALMYY165 pKa = 10.04TSTQPQILLLNNRR178 pKa = 11.84KK179 pKa = 9.94RR180 pKa = 11.84IVKK183 pKa = 10.15CKK185 pKa = 10.64ANNKK189 pKa = 6.93KK190 pKa = 7.91TKK192 pKa = 9.55PYY194 pKa = 9.32IRR196 pKa = 11.84LRR198 pKa = 11.84IPPPSQLKK206 pKa = 7.91TKK208 pKa = 9.47WYY210 pKa = 8.27FQRR213 pKa = 11.84DD214 pKa = 3.53IANLPLLLTIATACSLDD231 pKa = 3.41RR232 pKa = 11.84PYY234 pKa = 11.39LNSQSISNTVGFKK247 pKa = 9.26TLNYY251 pKa = 9.94EE252 pKa = 3.83AFLNRR257 pKa = 11.84GFKK260 pKa = 10.59NIPTTGYY267 pKa = 9.66FPKK270 pKa = 10.47PNTRR274 pKa = 11.84YY275 pKa = 10.08FITQTHH281 pKa = 5.14STDD284 pKa = 3.51PKK286 pKa = 9.94NVPIGSLIFLGNTNLLTLGTKK307 pKa = 9.45IQDD310 pKa = 3.57ACPSSTEE317 pKa = 4.15NEE319 pKa = 3.92WKK321 pKa = 10.34NQFEE325 pKa = 5.16KK326 pKa = 11.16YY327 pKa = 10.43LQTEE331 pKa = 4.55KK332 pKa = 10.88NWGNPFTPEE341 pKa = 3.91YY342 pKa = 10.66LDD344 pKa = 3.71EE345 pKa = 5.11DD346 pKa = 4.56NYY348 pKa = 11.32NIWITNKK355 pKa = 9.19TPQEE359 pKa = 3.82LKK361 pKa = 10.81AKK363 pKa = 10.1YY364 pKa = 9.86KK365 pKa = 11.02SNTDD369 pKa = 3.45KK370 pKa = 11.18LHH372 pKa = 7.29SSDD375 pKa = 4.78TDD377 pKa = 3.76TFWTLKK383 pKa = 10.1RR384 pKa = 11.84VPSVLDD390 pKa = 3.13CRR392 pKa = 11.84YY393 pKa = 10.83NPFRR397 pKa = 11.84DD398 pKa = 3.58DD399 pKa = 3.55NRR401 pKa = 11.84NEE403 pKa = 3.88VYY405 pKa = 10.52LLKK408 pKa = 9.76ITEE411 pKa = 4.15QNSYY415 pKa = 10.94DD416 pKa = 3.53YY417 pKa = 11.04HH418 pKa = 6.73EE419 pKa = 4.76PPKK422 pKa = 10.6PEE424 pKa = 5.76LIAKK428 pKa = 8.13DD429 pKa = 3.97LPLWACLWGYY439 pKa = 10.57IDD441 pKa = 4.06YY442 pKa = 10.5QKK444 pKa = 11.26KK445 pKa = 9.23AATYY449 pKa = 8.67TEE451 pKa = 4.3IDD453 pKa = 3.62TTCICVILSHH463 pKa = 6.98SLDD466 pKa = 3.86PTTHH470 pKa = 5.27TTFIPLDD477 pKa = 3.59EE478 pKa = 5.69DD479 pKa = 3.73FLKK482 pKa = 10.93GQSSFRR488 pKa = 11.84PEE490 pKa = 4.09HH491 pKa = 4.88QTTRR495 pKa = 11.84PDD497 pKa = 3.26EE498 pKa = 4.87LNWHH502 pKa = 6.7PKK504 pKa = 10.12VSFQIQSLNNIACTGPYY521 pKa = 7.83TAKK524 pKa = 10.59LPAGTSIEE532 pKa = 3.67GHH534 pKa = 4.97MQYY537 pKa = 10.94KK538 pKa = 9.31FHH540 pKa = 7.15FKK542 pKa = 10.65LGGSPPPMATLTNPEE557 pKa = 4.24NQPKK561 pKa = 9.78YY562 pKa = 9.77PNPDD566 pKa = 2.85NFLQTTSLQSPTMPIEE582 pKa = 4.45HH583 pKa = 6.64YY584 pKa = 10.44LYY586 pKa = 10.69NFDD589 pKa = 3.83EE590 pKa = 4.55RR591 pKa = 11.84RR592 pKa = 11.84HH593 pKa = 5.61ILTKK597 pKa = 10.51KK598 pKa = 9.95AIKK601 pKa = 10.32RR602 pKa = 11.84MQTDD606 pKa = 3.4KK607 pKa = 10.29EE608 pKa = 4.72TQSSLLQITDD618 pKa = 3.49SASTDD623 pKa = 3.04VPTPYY628 pKa = 10.36TEE630 pKa = 4.4TPTSEE635 pKa = 4.67SEE637 pKa = 4.05DD638 pKa = 3.62SEE640 pKa = 4.93KK641 pKa = 11.19EE642 pKa = 4.1EE643 pKa = 3.81EE644 pKa = 4.25TLQQRR649 pKa = 11.84LLKK652 pKa = 9.11QRR654 pKa = 11.84KK655 pKa = 6.38YY656 pKa = 10.73QKK658 pKa = 9.99QLQRR662 pKa = 11.84GINKK666 pKa = 9.67LLNRR670 pKa = 11.84LANIQQ675 pKa = 3.37
MM1 pKa = 7.79PWRR4 pKa = 11.84NYY6 pKa = 8.37YY7 pKa = 8.77YY8 pKa = 10.24RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84PRR15 pKa = 11.84RR16 pKa = 11.84FWRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84FRR23 pKa = 11.84TPIYY27 pKa = 9.42RR28 pKa = 11.84RR29 pKa = 11.84FWRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84YY35 pKa = 9.38RR36 pKa = 11.84VRR38 pKa = 11.84KK39 pKa = 8.82RR40 pKa = 11.84HH41 pKa = 5.88FKK43 pKa = 10.65RR44 pKa = 11.84KK45 pKa = 9.28LSKK48 pKa = 10.04LTIQEE53 pKa = 4.02WQPPSIRR60 pKa = 11.84KK61 pKa = 8.27LTIKK65 pKa = 10.42GYY67 pKa = 10.22YY68 pKa = 9.53PLFQSTRR75 pKa = 11.84DD76 pKa = 3.43RR77 pKa = 11.84LSNNMITYY85 pKa = 10.1LEE87 pKa = 4.88SIAPHH92 pKa = 5.51YY93 pKa = 10.59VPGGGGFSISCFSLNTLYY111 pKa = 10.67KK112 pKa = 10.53EE113 pKa = 4.07HH114 pKa = 7.13LKK116 pKa = 10.5LHH118 pKa = 5.54NWWTKK123 pKa = 10.15TNDD126 pKa = 3.19NYY128 pKa = 11.09PLIRR132 pKa = 11.84YY133 pKa = 7.46NGCTLKK139 pKa = 10.73LFKK142 pKa = 10.43SEE144 pKa = 3.83KK145 pKa = 9.1VDD147 pKa = 3.57YY148 pKa = 10.31IFYY151 pKa = 9.01YY152 pKa = 9.07QRR154 pKa = 11.84HH155 pKa = 5.38YY156 pKa = 10.69PMKK159 pKa = 10.46ASALMYY165 pKa = 10.04TSTQPQILLLNNRR178 pKa = 11.84KK179 pKa = 9.94RR180 pKa = 11.84IVKK183 pKa = 10.15CKK185 pKa = 10.64ANNKK189 pKa = 6.93KK190 pKa = 7.91TKK192 pKa = 9.55PYY194 pKa = 9.32IRR196 pKa = 11.84LRR198 pKa = 11.84IPPPSQLKK206 pKa = 7.91TKK208 pKa = 9.47WYY210 pKa = 8.27FQRR213 pKa = 11.84DD214 pKa = 3.53IANLPLLLTIATACSLDD231 pKa = 3.41RR232 pKa = 11.84PYY234 pKa = 11.39LNSQSISNTVGFKK247 pKa = 9.26TLNYY251 pKa = 9.94EE252 pKa = 3.83AFLNRR257 pKa = 11.84GFKK260 pKa = 10.59NIPTTGYY267 pKa = 9.66FPKK270 pKa = 10.47PNTRR274 pKa = 11.84YY275 pKa = 10.08FITQTHH281 pKa = 5.14STDD284 pKa = 3.51PKK286 pKa = 9.94NVPIGSLIFLGNTNLLTLGTKK307 pKa = 9.45IQDD310 pKa = 3.57ACPSSTEE317 pKa = 4.15NEE319 pKa = 3.92WKK321 pKa = 10.34NQFEE325 pKa = 5.16KK326 pKa = 11.16YY327 pKa = 10.43LQTEE331 pKa = 4.55KK332 pKa = 10.88NWGNPFTPEE341 pKa = 3.91YY342 pKa = 10.66LDD344 pKa = 3.71EE345 pKa = 5.11DD346 pKa = 4.56NYY348 pKa = 11.32NIWITNKK355 pKa = 9.19TPQEE359 pKa = 3.82LKK361 pKa = 10.81AKK363 pKa = 10.1YY364 pKa = 9.86KK365 pKa = 11.02SNTDD369 pKa = 3.45KK370 pKa = 11.18LHH372 pKa = 7.29SSDD375 pKa = 4.78TDD377 pKa = 3.76TFWTLKK383 pKa = 10.1RR384 pKa = 11.84VPSVLDD390 pKa = 3.13CRR392 pKa = 11.84YY393 pKa = 10.83NPFRR397 pKa = 11.84DD398 pKa = 3.58DD399 pKa = 3.55NRR401 pKa = 11.84NEE403 pKa = 3.88VYY405 pKa = 10.52LLKK408 pKa = 9.76ITEE411 pKa = 4.15QNSYY415 pKa = 10.94DD416 pKa = 3.53YY417 pKa = 11.04HH418 pKa = 6.73EE419 pKa = 4.76PPKK422 pKa = 10.6PEE424 pKa = 5.76LIAKK428 pKa = 8.13DD429 pKa = 3.97LPLWACLWGYY439 pKa = 10.57IDD441 pKa = 4.06YY442 pKa = 10.5QKK444 pKa = 11.26KK445 pKa = 9.23AATYY449 pKa = 8.67TEE451 pKa = 4.3IDD453 pKa = 3.62TTCICVILSHH463 pKa = 6.98SLDD466 pKa = 3.86PTTHH470 pKa = 5.27TTFIPLDD477 pKa = 3.59EE478 pKa = 5.69DD479 pKa = 3.73FLKK482 pKa = 10.93GQSSFRR488 pKa = 11.84PEE490 pKa = 4.09HH491 pKa = 4.88QTTRR495 pKa = 11.84PDD497 pKa = 3.26EE498 pKa = 4.87LNWHH502 pKa = 6.7PKK504 pKa = 10.12VSFQIQSLNNIACTGPYY521 pKa = 7.83TAKK524 pKa = 10.59LPAGTSIEE532 pKa = 3.67GHH534 pKa = 4.97MQYY537 pKa = 10.94KK538 pKa = 9.31FHH540 pKa = 7.15FKK542 pKa = 10.65LGGSPPPMATLTNPEE557 pKa = 4.24NQPKK561 pKa = 9.78YY562 pKa = 9.77PNPDD566 pKa = 2.85NFLQTTSLQSPTMPIEE582 pKa = 4.45HH583 pKa = 6.64YY584 pKa = 10.44LYY586 pKa = 10.69NFDD589 pKa = 3.83EE590 pKa = 4.55RR591 pKa = 11.84RR592 pKa = 11.84HH593 pKa = 5.61ILTKK597 pKa = 10.51KK598 pKa = 9.95AIKK601 pKa = 10.32RR602 pKa = 11.84MQTDD606 pKa = 3.4KK607 pKa = 10.29EE608 pKa = 4.72TQSSLLQITDD618 pKa = 3.49SASTDD623 pKa = 3.04VPTPYY628 pKa = 10.36TEE630 pKa = 4.4TPTSEE635 pKa = 4.67SEE637 pKa = 4.05DD638 pKa = 3.62SEE640 pKa = 4.93KK641 pKa = 11.19EE642 pKa = 4.1EE643 pKa = 3.81EE644 pKa = 4.25TLQQRR649 pKa = 11.84LLKK652 pKa = 9.11QRR654 pKa = 11.84KK655 pKa = 6.38YY656 pKa = 10.73QKK658 pKa = 9.99QLQRR662 pKa = 11.84GINKK666 pKa = 9.67LLNRR670 pKa = 11.84LANIQQ675 pKa = 3.37
Molecular weight: 79.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
774 |
99 |
675 |
387.0 |
45.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.101 ± 0.034 | 1.68 ± 0.645 |
5.039 ± 2.417 | 5.297 ± 1.812 |
4.393 ± 0.796 | 3.747 ± 2.07 |
2.584 ± 0.695 | 5.685 ± 0.303 |
8.01 ± 0.449 | 9.69 ± 0.286 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.163 ± 0.073 | 6.46 ± 0.673 |
7.106 ± 1.464 | 4.91 ± 0.415 |
6.46 ± 1.638 | 5.814 ± 1.329 |
9.69 ± 0.679 | 1.809 ± 0.101 |
1.938 ± 0.443 | 5.426 ± 2.109 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
