
Varroa destructor virus 3
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
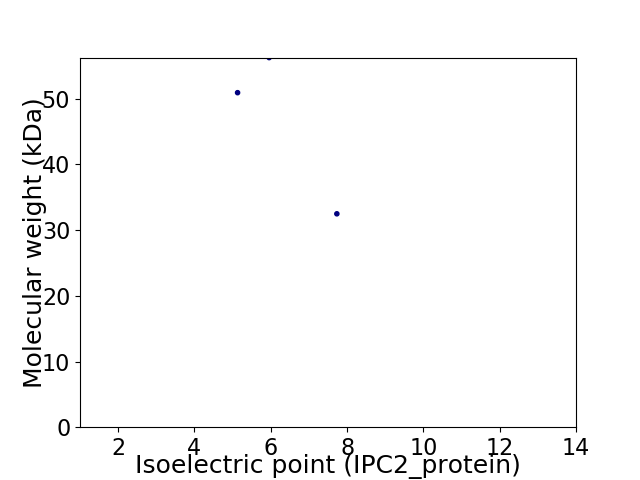
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1J0CQB9|A0A1J0CQB9_9VIRU RNA-directed RNA polymerase OS=Varroa destructor virus 3 OX=2056385 PE=4 SV=1
MM1 pKa = 7.31NCIEE5 pKa = 4.15LQYY8 pKa = 10.93TFDD11 pKa = 3.68LRR13 pKa = 11.84EE14 pKa = 3.59MRR16 pKa = 11.84FDD18 pKa = 5.4HH19 pKa = 7.31IICTLIWTMALIFLVVNGVLGLEE42 pKa = 3.89YY43 pKa = 10.22RR44 pKa = 11.84IKK46 pKa = 10.89NIMYY50 pKa = 10.94NMFLLDD56 pKa = 3.75APKK59 pKa = 8.5TTIYY63 pKa = 10.66EE64 pKa = 4.03DD65 pKa = 3.36TAMLEE70 pKa = 4.24SMKK73 pKa = 10.61VNPLVTAHH81 pKa = 7.64PDD83 pKa = 3.58TFPSCQLMVLTCNDD97 pKa = 2.9QGVWCRR103 pKa = 11.84AGNGFRR109 pKa = 11.84ISKK112 pKa = 8.41TIVVPTHH119 pKa = 6.51VIDD122 pKa = 4.29TSAFTAKK129 pKa = 9.89YY130 pKa = 9.79VRR132 pKa = 11.84LAVPEE137 pKa = 4.25GKK139 pKa = 10.1RR140 pKa = 11.84LGKK143 pKa = 9.23MVVVEE148 pKa = 4.32TPIKK152 pKa = 10.53DD153 pKa = 3.07ILTIGDD159 pKa = 4.94DD160 pKa = 3.12ISLINISEE168 pKa = 4.14IEE170 pKa = 4.12LNRR173 pKa = 11.84LGVTAARR180 pKa = 11.84IKK182 pKa = 10.04TLDD185 pKa = 3.45TKK187 pKa = 9.6QTSVMAMIYY196 pKa = 10.13SYY198 pKa = 11.78APLFKK203 pKa = 10.78ASVGKK208 pKa = 10.62ISPFDD213 pKa = 3.5GLAQQIVFSGSTIQGFSGALYY234 pKa = 10.23VAGKK238 pKa = 9.55NDD240 pKa = 3.66VYY242 pKa = 11.53GMHH245 pKa = 6.83TSGGAVNFGISASYY259 pKa = 10.24LQLLVLGTEE268 pKa = 4.39SNDD271 pKa = 3.94TIIKK275 pKa = 10.06SQDD278 pKa = 3.22NPLSRR283 pKa = 11.84LIKK286 pKa = 10.35RR287 pKa = 11.84FMEE290 pKa = 4.3LEE292 pKa = 4.31NGSSVTPEE300 pKa = 3.56SRR302 pKa = 11.84GGYY305 pKa = 9.64SSDD308 pKa = 3.19RR309 pKa = 11.84TLEE312 pKa = 4.03EE313 pKa = 5.39FLSDD317 pKa = 3.17WDD319 pKa = 3.79NKK321 pKa = 10.66KK322 pKa = 10.23KK323 pKa = 11.05GKK325 pKa = 9.75IGRR328 pKa = 11.84NWNDD332 pKa = 2.92SLGAYY337 pKa = 9.32VYY339 pKa = 10.41SIEE342 pKa = 3.88GRR344 pKa = 11.84FYY346 pKa = 11.21NITEE350 pKa = 5.13DD351 pKa = 3.24IDD353 pKa = 3.73NYY355 pKa = 10.04AQIRR359 pKa = 11.84FPHH362 pKa = 6.44LFEE365 pKa = 6.01DD366 pKa = 4.98ADD368 pKa = 3.98FEE370 pKa = 5.18YY371 pKa = 9.18DD372 pKa = 3.3THH374 pKa = 8.38DD375 pKa = 3.43EE376 pKa = 4.34NGDD379 pKa = 3.05KK380 pKa = 10.41RR381 pKa = 11.84VRR383 pKa = 11.84HH384 pKa = 5.62NKK386 pKa = 8.65FSKK389 pKa = 9.67SKK391 pKa = 10.29RR392 pKa = 11.84YY393 pKa = 10.0ANPIEE398 pKa = 4.97DD399 pKa = 4.98EE400 pKa = 4.37IPVEE404 pKa = 4.5DD405 pKa = 3.69IYY407 pKa = 11.43EE408 pKa = 4.13EE409 pKa = 4.18EE410 pKa = 3.84EE411 pKa = 4.23EE412 pKa = 4.23YY413 pKa = 10.81RR414 pKa = 11.84LEE416 pKa = 3.61EE417 pKa = 3.88RR418 pKa = 11.84RR419 pKa = 11.84RR420 pKa = 11.84KK421 pKa = 9.7KK422 pKa = 10.84VEE424 pKa = 3.26FDD426 pKa = 3.12EE427 pKa = 5.7FVRR430 pKa = 11.84NMNWPVVLGGLMNQLLSEE448 pKa = 4.27
MM1 pKa = 7.31NCIEE5 pKa = 4.15LQYY8 pKa = 10.93TFDD11 pKa = 3.68LRR13 pKa = 11.84EE14 pKa = 3.59MRR16 pKa = 11.84FDD18 pKa = 5.4HH19 pKa = 7.31IICTLIWTMALIFLVVNGVLGLEE42 pKa = 3.89YY43 pKa = 10.22RR44 pKa = 11.84IKK46 pKa = 10.89NIMYY50 pKa = 10.94NMFLLDD56 pKa = 3.75APKK59 pKa = 8.5TTIYY63 pKa = 10.66EE64 pKa = 4.03DD65 pKa = 3.36TAMLEE70 pKa = 4.24SMKK73 pKa = 10.61VNPLVTAHH81 pKa = 7.64PDD83 pKa = 3.58TFPSCQLMVLTCNDD97 pKa = 2.9QGVWCRR103 pKa = 11.84AGNGFRR109 pKa = 11.84ISKK112 pKa = 8.41TIVVPTHH119 pKa = 6.51VIDD122 pKa = 4.29TSAFTAKK129 pKa = 9.89YY130 pKa = 9.79VRR132 pKa = 11.84LAVPEE137 pKa = 4.25GKK139 pKa = 10.1RR140 pKa = 11.84LGKK143 pKa = 9.23MVVVEE148 pKa = 4.32TPIKK152 pKa = 10.53DD153 pKa = 3.07ILTIGDD159 pKa = 4.94DD160 pKa = 3.12ISLINISEE168 pKa = 4.14IEE170 pKa = 4.12LNRR173 pKa = 11.84LGVTAARR180 pKa = 11.84IKK182 pKa = 10.04TLDD185 pKa = 3.45TKK187 pKa = 9.6QTSVMAMIYY196 pKa = 10.13SYY198 pKa = 11.78APLFKK203 pKa = 10.78ASVGKK208 pKa = 10.62ISPFDD213 pKa = 3.5GLAQQIVFSGSTIQGFSGALYY234 pKa = 10.23VAGKK238 pKa = 9.55NDD240 pKa = 3.66VYY242 pKa = 11.53GMHH245 pKa = 6.83TSGGAVNFGISASYY259 pKa = 10.24LQLLVLGTEE268 pKa = 4.39SNDD271 pKa = 3.94TIIKK275 pKa = 10.06SQDD278 pKa = 3.22NPLSRR283 pKa = 11.84LIKK286 pKa = 10.35RR287 pKa = 11.84FMEE290 pKa = 4.3LEE292 pKa = 4.31NGSSVTPEE300 pKa = 3.56SRR302 pKa = 11.84GGYY305 pKa = 9.64SSDD308 pKa = 3.19RR309 pKa = 11.84TLEE312 pKa = 4.03EE313 pKa = 5.39FLSDD317 pKa = 3.17WDD319 pKa = 3.79NKK321 pKa = 10.66KK322 pKa = 10.23KK323 pKa = 11.05GKK325 pKa = 9.75IGRR328 pKa = 11.84NWNDD332 pKa = 2.92SLGAYY337 pKa = 9.32VYY339 pKa = 10.41SIEE342 pKa = 3.88GRR344 pKa = 11.84FYY346 pKa = 11.21NITEE350 pKa = 5.13DD351 pKa = 3.24IDD353 pKa = 3.73NYY355 pKa = 10.04AQIRR359 pKa = 11.84FPHH362 pKa = 6.44LFEE365 pKa = 6.01DD366 pKa = 4.98ADD368 pKa = 3.98FEE370 pKa = 5.18YY371 pKa = 9.18DD372 pKa = 3.3THH374 pKa = 8.38DD375 pKa = 3.43EE376 pKa = 4.34NGDD379 pKa = 3.05KK380 pKa = 10.41RR381 pKa = 11.84VRR383 pKa = 11.84HH384 pKa = 5.62NKK386 pKa = 8.65FSKK389 pKa = 9.67SKK391 pKa = 10.29RR392 pKa = 11.84YY393 pKa = 10.0ANPIEE398 pKa = 4.97DD399 pKa = 4.98EE400 pKa = 4.37IPVEE404 pKa = 4.5DD405 pKa = 3.69IYY407 pKa = 11.43EE408 pKa = 4.13EE409 pKa = 4.18EE410 pKa = 3.84EE411 pKa = 4.23EE412 pKa = 4.23YY413 pKa = 10.81RR414 pKa = 11.84LEE416 pKa = 3.61EE417 pKa = 3.88RR418 pKa = 11.84RR419 pKa = 11.84RR420 pKa = 11.84KK421 pKa = 9.7KK422 pKa = 10.84VEE424 pKa = 3.26FDD426 pKa = 3.12EE427 pKa = 5.7FVRR430 pKa = 11.84NMNWPVVLGGLMNQLLSEE448 pKa = 4.27
Molecular weight: 50.92 kDa
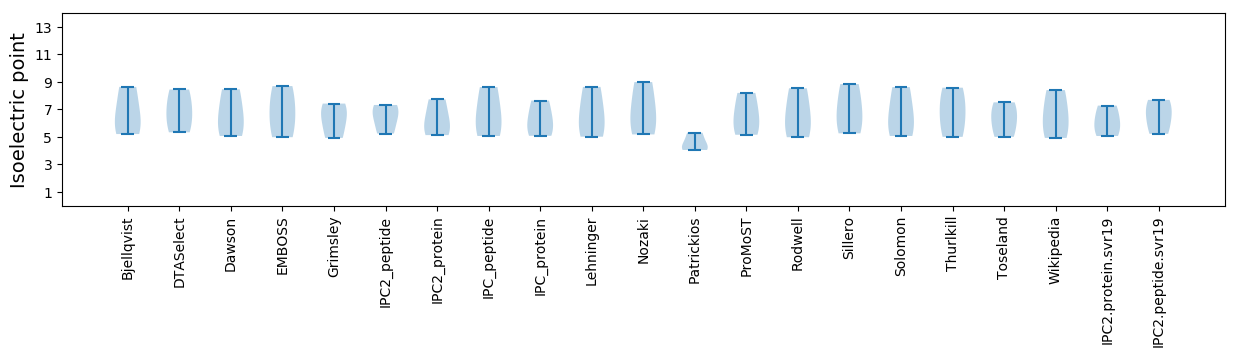
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1J0CQC2|A0A1J0CQC2_9VIRU ORF2 OS=Varroa destructor virus 3 OX=2056385 PE=4 SV=1
MM1 pKa = 7.37KK2 pKa = 10.43RR3 pKa = 11.84LLIQEE8 pKa = 4.69ANANPNGGYY17 pKa = 9.94KK18 pKa = 10.18DD19 pKa = 4.05DD20 pKa = 4.28VLMEE24 pKa = 5.11PIFHH28 pKa = 7.09GDD30 pKa = 3.18SEE32 pKa = 5.05AEE34 pKa = 4.05LLADD38 pKa = 4.19YY39 pKa = 10.67NQMVAPMLVEE49 pKa = 5.4DD50 pKa = 4.59EE51 pKa = 4.96LDD53 pKa = 3.58LLRR56 pKa = 11.84EE57 pKa = 4.28TVNKK61 pKa = 10.62SEE63 pKa = 3.96VSYY66 pKa = 11.56EE67 pKa = 3.89EE68 pKa = 4.0VQSIRR73 pKa = 11.84SICNRR78 pKa = 11.84LTQLGKK84 pKa = 10.56FSNQLLSTIDD94 pKa = 4.11KK95 pKa = 10.5IRR97 pKa = 11.84ADD99 pKa = 3.33KK100 pKa = 10.58KK101 pKa = 10.14KK102 pKa = 10.74RR103 pKa = 11.84EE104 pKa = 4.11AEE106 pKa = 3.69EE107 pKa = 3.64RR108 pKa = 11.84ARR110 pKa = 11.84QIEE113 pKa = 4.42MEE115 pKa = 4.02LKK117 pKa = 8.84MRR119 pKa = 11.84KK120 pKa = 9.21KK121 pKa = 10.71EE122 pKa = 3.97EE123 pKa = 3.89LKK125 pKa = 10.67KK126 pKa = 10.48RR127 pKa = 11.84AIEE130 pKa = 4.04MEE132 pKa = 4.13EE133 pKa = 3.67SRR135 pKa = 11.84ARR137 pKa = 11.84QEE139 pKa = 4.19SEE141 pKa = 4.0AKK143 pKa = 9.13EE144 pKa = 4.22KK145 pKa = 10.16EE146 pKa = 4.47VKK148 pKa = 10.25RR149 pKa = 11.84SAMCLRR155 pKa = 11.84VRR157 pKa = 11.84EE158 pKa = 4.2LFEE161 pKa = 4.39SLTEE165 pKa = 3.96EE166 pKa = 4.3EE167 pKa = 4.05IKK169 pKa = 10.73EE170 pKa = 3.89FSRR173 pKa = 11.84LVVEE177 pKa = 4.99RR178 pKa = 11.84NTKK181 pKa = 9.84RR182 pKa = 11.84ALARR186 pKa = 11.84KK187 pKa = 8.95SEE189 pKa = 4.11KK190 pKa = 10.16EE191 pKa = 3.75KK192 pKa = 10.25MRR194 pKa = 11.84QEE196 pKa = 4.27VISAIDD202 pKa = 3.47RR203 pKa = 11.84PEE205 pKa = 3.88EE206 pKa = 4.13KK207 pKa = 9.88SDD209 pKa = 3.41KK210 pKa = 10.85KK211 pKa = 10.77IVLEE215 pKa = 4.14GTKK218 pKa = 10.73LKK220 pKa = 9.48DD221 pKa = 3.24TKK223 pKa = 11.15SSVKK227 pKa = 10.26EE228 pKa = 4.36DD229 pKa = 3.01IRR231 pKa = 11.84PPPKK235 pKa = 10.74GMIRR239 pKa = 11.84CPSCIIRR246 pKa = 11.84KK247 pKa = 9.72AKK249 pKa = 10.23DD250 pKa = 3.4RR251 pKa = 11.84PCFSNKK257 pKa = 9.24PDD259 pKa = 3.67FFKK262 pKa = 11.01HH263 pKa = 4.91MEE265 pKa = 4.25EE266 pKa = 4.01VHH268 pKa = 5.33LTRR271 pKa = 11.84LEE273 pKa = 4.12AAPSGNN279 pKa = 3.3
MM1 pKa = 7.37KK2 pKa = 10.43RR3 pKa = 11.84LLIQEE8 pKa = 4.69ANANPNGGYY17 pKa = 9.94KK18 pKa = 10.18DD19 pKa = 4.05DD20 pKa = 4.28VLMEE24 pKa = 5.11PIFHH28 pKa = 7.09GDD30 pKa = 3.18SEE32 pKa = 5.05AEE34 pKa = 4.05LLADD38 pKa = 4.19YY39 pKa = 10.67NQMVAPMLVEE49 pKa = 5.4DD50 pKa = 4.59EE51 pKa = 4.96LDD53 pKa = 3.58LLRR56 pKa = 11.84EE57 pKa = 4.28TVNKK61 pKa = 10.62SEE63 pKa = 3.96VSYY66 pKa = 11.56EE67 pKa = 3.89EE68 pKa = 4.0VQSIRR73 pKa = 11.84SICNRR78 pKa = 11.84LTQLGKK84 pKa = 10.56FSNQLLSTIDD94 pKa = 4.11KK95 pKa = 10.5IRR97 pKa = 11.84ADD99 pKa = 3.33KK100 pKa = 10.58KK101 pKa = 10.14KK102 pKa = 10.74RR103 pKa = 11.84EE104 pKa = 4.11AEE106 pKa = 3.69EE107 pKa = 3.64RR108 pKa = 11.84ARR110 pKa = 11.84QIEE113 pKa = 4.42MEE115 pKa = 4.02LKK117 pKa = 8.84MRR119 pKa = 11.84KK120 pKa = 9.21KK121 pKa = 10.71EE122 pKa = 3.97EE123 pKa = 3.89LKK125 pKa = 10.67KK126 pKa = 10.48RR127 pKa = 11.84AIEE130 pKa = 4.04MEE132 pKa = 4.13EE133 pKa = 3.67SRR135 pKa = 11.84ARR137 pKa = 11.84QEE139 pKa = 4.19SEE141 pKa = 4.0AKK143 pKa = 9.13EE144 pKa = 4.22KK145 pKa = 10.16EE146 pKa = 4.47VKK148 pKa = 10.25RR149 pKa = 11.84SAMCLRR155 pKa = 11.84VRR157 pKa = 11.84EE158 pKa = 4.2LFEE161 pKa = 4.39SLTEE165 pKa = 3.96EE166 pKa = 4.3EE167 pKa = 4.05IKK169 pKa = 10.73EE170 pKa = 3.89FSRR173 pKa = 11.84LVVEE177 pKa = 4.99RR178 pKa = 11.84NTKK181 pKa = 9.84RR182 pKa = 11.84ALARR186 pKa = 11.84KK187 pKa = 8.95SEE189 pKa = 4.11KK190 pKa = 10.16EE191 pKa = 3.75KK192 pKa = 10.25MRR194 pKa = 11.84QEE196 pKa = 4.27VISAIDD202 pKa = 3.47RR203 pKa = 11.84PEE205 pKa = 3.88EE206 pKa = 4.13KK207 pKa = 9.88SDD209 pKa = 3.41KK210 pKa = 10.85KK211 pKa = 10.77IVLEE215 pKa = 4.14GTKK218 pKa = 10.73LKK220 pKa = 9.48DD221 pKa = 3.24TKK223 pKa = 11.15SSVKK227 pKa = 10.26EE228 pKa = 4.36DD229 pKa = 3.01IRR231 pKa = 11.84PPPKK235 pKa = 10.74GMIRR239 pKa = 11.84CPSCIIRR246 pKa = 11.84KK247 pKa = 9.72AKK249 pKa = 10.23DD250 pKa = 3.4RR251 pKa = 11.84PCFSNKK257 pKa = 9.24PDD259 pKa = 3.67FFKK262 pKa = 11.01HH263 pKa = 4.91MEE265 pKa = 4.25EE266 pKa = 4.01VHH268 pKa = 5.33LTRR271 pKa = 11.84LEE273 pKa = 4.12AAPSGNN279 pKa = 3.3
Molecular weight: 32.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1220 |
279 |
493 |
406.7 |
46.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.393 ± 0.802 | 1.639 ± 0.265 |
5.902 ± 0.31 | 8.934 ± 1.713 |
4.344 ± 0.557 | 5.656 ± 0.935 |
2.049 ± 0.516 | 5.738 ± 1.134 |
7.049 ± 1.419 | 9.344 ± 0.521 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.443 ± 0.152 | 4.426 ± 0.551 |
4.098 ± 0.439 | 2.459 ± 0.138 |
6.639 ± 0.836 | 6.311 ± 0.435 |
4.262 ± 0.842 | 6.803 ± 0.579 |
1.557 ± 0.681 | 2.951 ± 0.682 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
