
Octadecabacter sp. SW4
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Octadecabacter; unclassified Octadecabacter
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
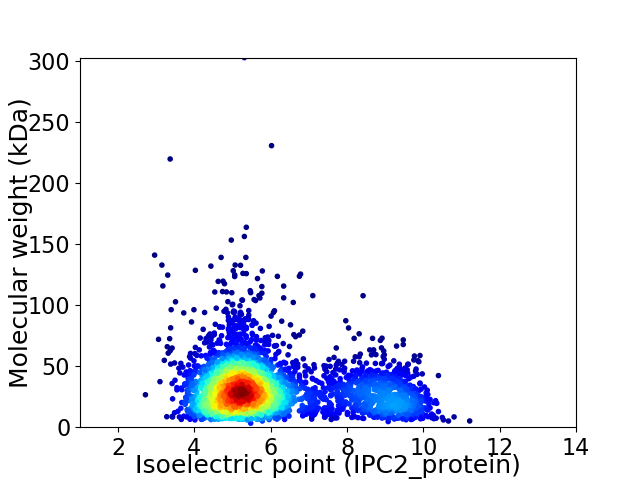
Virtual 2D-PAGE plot for 3350 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
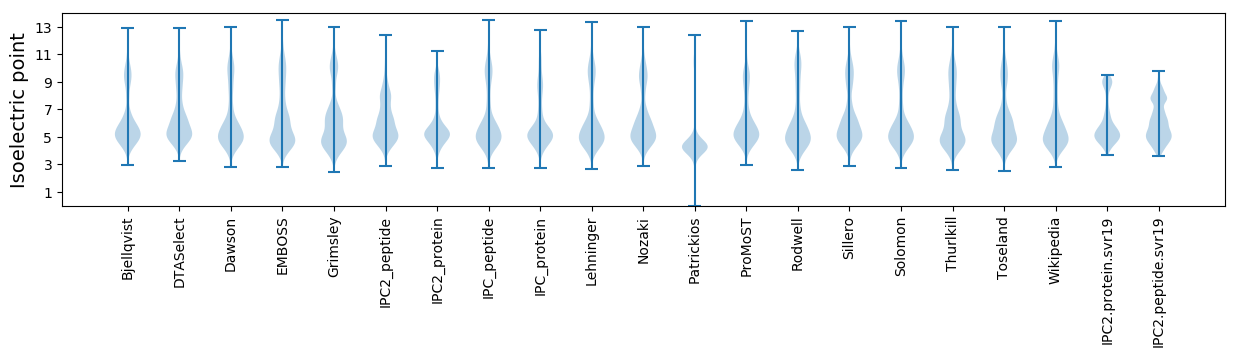
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B9ENM2|A0A5B9ENM2_9RHOB Molybdenum import ATP-binding protein ModC OS=Octadecabacter sp. SW4 OX=2602067 GN=modC PE=3 SV=1
MM1 pKa = 6.7QAAAGFRR8 pKa = 11.84RR9 pKa = 11.84VWPSLLIKK17 pKa = 10.29GIGMTAITTNVQGYY31 pKa = 5.82GTQWFSAPGDD41 pKa = 3.83NLHH44 pKa = 6.24VLSTGLLGSWDD55 pKa = 4.03TLTALASHH63 pKa = 7.45DD64 pKa = 3.84NLRR67 pKa = 11.84VDD69 pKa = 4.55NSGQMWSMASVLEE82 pKa = 4.1VRR84 pKa = 11.84GSDD87 pKa = 3.31VRR89 pKa = 11.84TANYY93 pKa = 10.01EE94 pKa = 3.92GGLMQSEE101 pKa = 4.57SDD103 pKa = 3.57SHH105 pKa = 7.83LDD107 pKa = 3.21AVIRR111 pKa = 11.84YY112 pKa = 9.02KK113 pKa = 11.21GEE115 pKa = 3.83AVGGQIFNFGTINAYY130 pKa = 8.39WAVGVEE136 pKa = 3.97IGTTGTTFDD145 pKa = 3.59NRR147 pKa = 11.84GLIDD151 pKa = 4.1AGSALRR157 pKa = 11.84MGLDD161 pKa = 3.48GADD164 pKa = 3.45GQVVRR169 pKa = 11.84NFGTMISAGNASAAGDD185 pKa = 3.53ATYY188 pKa = 11.33GNGVYY193 pKa = 10.18VEE195 pKa = 4.46GSNNLFYY202 pKa = 11.11NWTGSTLTVTSGGAGVAFGGAGTGNSVKK230 pKa = 10.57NAGTITSDD238 pKa = 3.41TGWGVDD244 pKa = 3.74LSGLAVGVSADD255 pKa = 3.74VINTRR260 pKa = 11.84GGVIAGGAGAVRR272 pKa = 11.84GNSAEE277 pKa = 3.89NVIYY281 pKa = 10.75NSGIMNGDD289 pKa = 2.77VWLYY293 pKa = 11.28GGDD296 pKa = 4.19DD297 pKa = 3.91YY298 pKa = 12.11VSSGTYY304 pKa = 10.02AQFNGDD310 pKa = 3.98LYY312 pKa = 11.27AGNGNDD318 pKa = 3.41TLYY321 pKa = 11.33GSGGDD326 pKa = 3.61DD327 pKa = 2.92SFYY330 pKa = 11.3GQGQNDD336 pKa = 4.73EE337 pKa = 3.88IWGFDD342 pKa = 3.2GDD344 pKa = 4.51DD345 pKa = 3.83FLFGGAGDD353 pKa = 4.29DD354 pKa = 4.17VIEE357 pKa = 5.03GGDD360 pKa = 3.51GADD363 pKa = 3.56YY364 pKa = 11.23LRR366 pKa = 11.84GQGGSDD372 pKa = 3.28TLNGGEE378 pKa = 4.78GDD380 pKa = 3.72DD381 pKa = 5.17DD382 pKa = 4.31IYY384 pKa = 11.52GGVGADD390 pKa = 2.57IFVFEE395 pKa = 4.41QNGNFDD401 pKa = 5.54IIWDD405 pKa = 3.91FQNNVDD411 pKa = 5.46QIDD414 pKa = 4.06LSDD417 pKa = 4.52LGITSYY423 pKa = 11.29SALAAVMTTVGADD436 pKa = 3.39VQIDD440 pKa = 4.18LSSLSGVYY448 pKa = 10.64GDD450 pKa = 4.2IIEE453 pKa = 4.57LRR455 pKa = 11.84GSVTIGDD462 pKa = 4.51LDD464 pKa = 4.63AGDD467 pKa = 5.24FILL470 pKa = 5.92
MM1 pKa = 6.7QAAAGFRR8 pKa = 11.84RR9 pKa = 11.84VWPSLLIKK17 pKa = 10.29GIGMTAITTNVQGYY31 pKa = 5.82GTQWFSAPGDD41 pKa = 3.83NLHH44 pKa = 6.24VLSTGLLGSWDD55 pKa = 4.03TLTALASHH63 pKa = 7.45DD64 pKa = 3.84NLRR67 pKa = 11.84VDD69 pKa = 4.55NSGQMWSMASVLEE82 pKa = 4.1VRR84 pKa = 11.84GSDD87 pKa = 3.31VRR89 pKa = 11.84TANYY93 pKa = 10.01EE94 pKa = 3.92GGLMQSEE101 pKa = 4.57SDD103 pKa = 3.57SHH105 pKa = 7.83LDD107 pKa = 3.21AVIRR111 pKa = 11.84YY112 pKa = 9.02KK113 pKa = 11.21GEE115 pKa = 3.83AVGGQIFNFGTINAYY130 pKa = 8.39WAVGVEE136 pKa = 3.97IGTTGTTFDD145 pKa = 3.59NRR147 pKa = 11.84GLIDD151 pKa = 4.1AGSALRR157 pKa = 11.84MGLDD161 pKa = 3.48GADD164 pKa = 3.45GQVVRR169 pKa = 11.84NFGTMISAGNASAAGDD185 pKa = 3.53ATYY188 pKa = 11.33GNGVYY193 pKa = 10.18VEE195 pKa = 4.46GSNNLFYY202 pKa = 11.11NWTGSTLTVTSGGAGVAFGGAGTGNSVKK230 pKa = 10.57NAGTITSDD238 pKa = 3.41TGWGVDD244 pKa = 3.74LSGLAVGVSADD255 pKa = 3.74VINTRR260 pKa = 11.84GGVIAGGAGAVRR272 pKa = 11.84GNSAEE277 pKa = 3.89NVIYY281 pKa = 10.75NSGIMNGDD289 pKa = 2.77VWLYY293 pKa = 11.28GGDD296 pKa = 4.19DD297 pKa = 3.91YY298 pKa = 12.11VSSGTYY304 pKa = 10.02AQFNGDD310 pKa = 3.98LYY312 pKa = 11.27AGNGNDD318 pKa = 3.41TLYY321 pKa = 11.33GSGGDD326 pKa = 3.61DD327 pKa = 2.92SFYY330 pKa = 11.3GQGQNDD336 pKa = 4.73EE337 pKa = 3.88IWGFDD342 pKa = 3.2GDD344 pKa = 4.51DD345 pKa = 3.83FLFGGAGDD353 pKa = 4.29DD354 pKa = 4.17VIEE357 pKa = 5.03GGDD360 pKa = 3.51GADD363 pKa = 3.56YY364 pKa = 11.23LRR366 pKa = 11.84GQGGSDD372 pKa = 3.28TLNGGEE378 pKa = 4.78GDD380 pKa = 3.72DD381 pKa = 5.17DD382 pKa = 4.31IYY384 pKa = 11.52GGVGADD390 pKa = 2.57IFVFEE395 pKa = 4.41QNGNFDD401 pKa = 5.54IIWDD405 pKa = 3.91FQNNVDD411 pKa = 5.46QIDD414 pKa = 4.06LSDD417 pKa = 4.52LGITSYY423 pKa = 11.29SALAAVMTTVGADD436 pKa = 3.39VQIDD440 pKa = 4.18LSSLSGVYY448 pKa = 10.64GDD450 pKa = 4.2IIEE453 pKa = 4.57LRR455 pKa = 11.84GSVTIGDD462 pKa = 4.51LDD464 pKa = 4.63AGDD467 pKa = 5.24FILL470 pKa = 5.92
Molecular weight: 48.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B9ENQ5|A0A5B9ENQ5_9RHOB Cell division protein FtsQ OS=Octadecabacter sp. SW4 OX=2602067 GN=ftsQ PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.56GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.56GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1034289 |
32 |
2756 |
308.7 |
33.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.518 ± 0.058 | 0.882 ± 0.013 |
6.598 ± 0.044 | 5.205 ± 0.042 |
3.824 ± 0.03 | 8.757 ± 0.048 |
2.03 ± 0.023 | 5.481 ± 0.029 |
3.072 ± 0.038 | 9.767 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.907 ± 0.02 | 2.693 ± 0.026 |
4.882 ± 0.028 | 3.294 ± 0.02 |
6.42 ± 0.038 | 4.854 ± 0.025 |
5.755 ± 0.029 | 7.386 ± 0.037 |
1.414 ± 0.02 | 2.261 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
