
Jatrophihabitans endophyticus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Jatrophihabitantales; Jatrophihabitantaceae; Jatrophihabitans
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
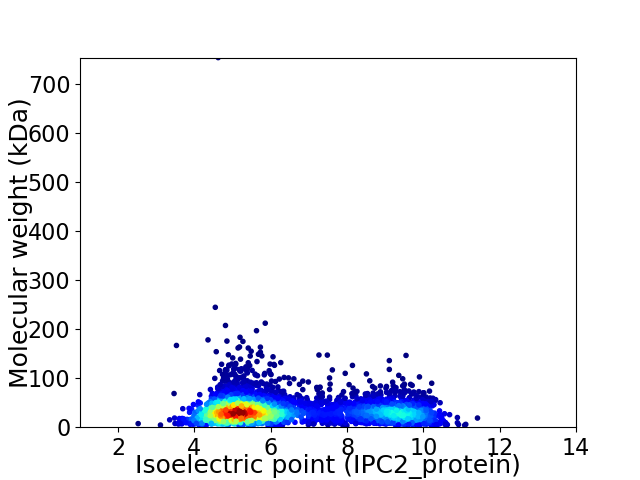
Virtual 2D-PAGE plot for 4181 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M5C0L1|A0A1M5C0L1_9ACTN Uncharacterized protein OS=Jatrophihabitans endophyticus OX=1206085 GN=SAMN05443575_0061 PE=4 SV=1
MM1 pKa = 7.38NIWLKK6 pKa = 10.84RR7 pKa = 11.84SLQMGLLGAGVWLVGTGIAEE27 pKa = 4.74ADD29 pKa = 3.74TAPAVPPSQVSLAHH43 pKa = 6.19VDD45 pKa = 2.83VDD47 pKa = 4.05VPVTVGHH54 pKa = 5.6NHH56 pKa = 3.79VTVANGSGDD65 pKa = 3.6ASAHH69 pKa = 6.22SDD71 pKa = 3.14TSAKK75 pKa = 10.32AGSSSGSSSRR85 pKa = 11.84SSVVDD90 pKa = 3.37VSVHH94 pKa = 4.96VPVTVSDD101 pKa = 3.54NDD103 pKa = 3.49MAVADD108 pKa = 4.66GSNSGGSNSGGSNSGGSNSGGSNSGGSNSGGSKK141 pKa = 9.55DD142 pKa = 3.48TDD144 pKa = 3.29NGGSAKK150 pKa = 10.43SGADD154 pKa = 3.08NGGDD158 pKa = 3.41EE159 pKa = 5.38KK160 pKa = 11.45GGADD164 pKa = 3.23AKK166 pKa = 10.6QRR168 pKa = 11.84SLVSVPVSAPITVAGNAVGVLGDD191 pKa = 4.5ADD193 pKa = 4.02SSSTGAGSAGSGGASASDD211 pKa = 3.65GAVEE215 pKa = 4.42VPVNAPVTVCGNGVGVLGEE234 pKa = 4.22GTADD238 pKa = 3.6CGAGSTSSGASGDD251 pKa = 3.58GSLVEE256 pKa = 4.43VPVNAPVTVCGNGVGVAGDD275 pKa = 3.68AAAGCGAGSAGSTSTGSGGASASDD299 pKa = 3.65GAVEE303 pKa = 4.42VPVNAPVTVCGNGVGVAGDD322 pKa = 3.68AAAGCGAGSAGSTSTGSGGASASDD346 pKa = 3.75GAVEE350 pKa = 4.42APVNAPVTVCGNGVGVAGDD369 pKa = 3.68AAAGCGAGSAGSTSTGSGGASASDD393 pKa = 3.65GAVEE397 pKa = 4.42VPVNAPVTVCGNGVGALGDD416 pKa = 3.85ASADD420 pKa = 3.78CASGSTGGATTGGSDD435 pKa = 3.44GSTGGDD441 pKa = 3.38DD442 pKa = 3.73GSVAEE447 pKa = 4.7VPVNAPVTVCGNGVGVAGDD466 pKa = 3.68AAAGCGAGSTAGSSTGSGASTGNDD490 pKa = 2.84GGVAEE495 pKa = 4.85VPVNAPVTVCGNGVGALGDD514 pKa = 3.76ASADD518 pKa = 3.78CTSGSTGGATTGGSDD533 pKa = 3.44GSTGGDD539 pKa = 3.38DD540 pKa = 3.73GSVAEE545 pKa = 4.7VPVNAPVTVCGNGVGALGDD564 pKa = 3.76ASADD568 pKa = 3.78CTSGSTGGATTGGSDD583 pKa = 3.44GSTGGDD589 pKa = 3.41DD590 pKa = 4.12TIVDD594 pKa = 4.01VPVEE598 pKa = 4.26VPVTVCGNAVGSADD612 pKa = 3.92SSCSTTTTTGGGTDD626 pKa = 3.3GGTDD630 pKa = 3.29GGTVTVPVDD639 pKa = 3.63TPVTVCGNGVGLLGTASASCSTGGDD664 pKa = 3.4TPGTNDD670 pKa = 4.63PGTNDD675 pKa = 4.35PGSNDD680 pKa = 4.02PGTNDD685 pKa = 4.44PGSNDD690 pKa = 3.87PGSDD694 pKa = 3.35NPTVTDD700 pKa = 4.0PVNAGSDD707 pKa = 3.73TTVEE711 pKa = 4.39AATSTPDD718 pKa = 3.01SHH720 pKa = 9.44SSDD723 pKa = 3.33TVGGGRR729 pKa = 11.84QLAQTGADD737 pKa = 3.28TRR739 pKa = 11.84GVLVGAVLLMLLGCVLGFAGRR760 pKa = 11.84RR761 pKa = 11.84RR762 pKa = 11.84TNGG765 pKa = 2.91
MM1 pKa = 7.38NIWLKK6 pKa = 10.84RR7 pKa = 11.84SLQMGLLGAGVWLVGTGIAEE27 pKa = 4.74ADD29 pKa = 3.74TAPAVPPSQVSLAHH43 pKa = 6.19VDD45 pKa = 2.83VDD47 pKa = 4.05VPVTVGHH54 pKa = 5.6NHH56 pKa = 3.79VTVANGSGDD65 pKa = 3.6ASAHH69 pKa = 6.22SDD71 pKa = 3.14TSAKK75 pKa = 10.32AGSSSGSSSRR85 pKa = 11.84SSVVDD90 pKa = 3.37VSVHH94 pKa = 4.96VPVTVSDD101 pKa = 3.54NDD103 pKa = 3.49MAVADD108 pKa = 4.66GSNSGGSNSGGSNSGGSNSGGSNSGGSNSGGSKK141 pKa = 9.55DD142 pKa = 3.48TDD144 pKa = 3.29NGGSAKK150 pKa = 10.43SGADD154 pKa = 3.08NGGDD158 pKa = 3.41EE159 pKa = 5.38KK160 pKa = 11.45GGADD164 pKa = 3.23AKK166 pKa = 10.6QRR168 pKa = 11.84SLVSVPVSAPITVAGNAVGVLGDD191 pKa = 4.5ADD193 pKa = 4.02SSSTGAGSAGSGGASASDD211 pKa = 3.65GAVEE215 pKa = 4.42VPVNAPVTVCGNGVGVLGEE234 pKa = 4.22GTADD238 pKa = 3.6CGAGSTSSGASGDD251 pKa = 3.58GSLVEE256 pKa = 4.43VPVNAPVTVCGNGVGVAGDD275 pKa = 3.68AAAGCGAGSAGSTSTGSGGASASDD299 pKa = 3.65GAVEE303 pKa = 4.42VPVNAPVTVCGNGVGVAGDD322 pKa = 3.68AAAGCGAGSAGSTSTGSGGASASDD346 pKa = 3.75GAVEE350 pKa = 4.42APVNAPVTVCGNGVGVAGDD369 pKa = 3.68AAAGCGAGSAGSTSTGSGGASASDD393 pKa = 3.65GAVEE397 pKa = 4.42VPVNAPVTVCGNGVGALGDD416 pKa = 3.85ASADD420 pKa = 3.78CASGSTGGATTGGSDD435 pKa = 3.44GSTGGDD441 pKa = 3.38DD442 pKa = 3.73GSVAEE447 pKa = 4.7VPVNAPVTVCGNGVGVAGDD466 pKa = 3.68AAAGCGAGSTAGSSTGSGASTGNDD490 pKa = 2.84GGVAEE495 pKa = 4.85VPVNAPVTVCGNGVGALGDD514 pKa = 3.76ASADD518 pKa = 3.78CTSGSTGGATTGGSDD533 pKa = 3.44GSTGGDD539 pKa = 3.38DD540 pKa = 3.73GSVAEE545 pKa = 4.7VPVNAPVTVCGNGVGALGDD564 pKa = 3.76ASADD568 pKa = 3.78CTSGSTGGATTGGSDD583 pKa = 3.44GSTGGDD589 pKa = 3.41DD590 pKa = 4.12TIVDD594 pKa = 4.01VPVEE598 pKa = 4.26VPVTVCGNAVGSADD612 pKa = 3.92SSCSTTTTTGGGTDD626 pKa = 3.3GGTDD630 pKa = 3.29GGTVTVPVDD639 pKa = 3.63TPVTVCGNGVGLLGTASASCSTGGDD664 pKa = 3.4TPGTNDD670 pKa = 4.63PGTNDD675 pKa = 4.35PGSNDD680 pKa = 4.02PGTNDD685 pKa = 4.44PGSNDD690 pKa = 3.87PGSDD694 pKa = 3.35NPTVTDD700 pKa = 4.0PVNAGSDD707 pKa = 3.73TTVEE711 pKa = 4.39AATSTPDD718 pKa = 3.01SHH720 pKa = 9.44SSDD723 pKa = 3.33TVGGGRR729 pKa = 11.84QLAQTGADD737 pKa = 3.28TRR739 pKa = 11.84GVLVGAVLLMLLGCVLGFAGRR760 pKa = 11.84RR761 pKa = 11.84RR762 pKa = 11.84TNGG765 pKa = 2.91
Molecular weight: 68.58 kDa
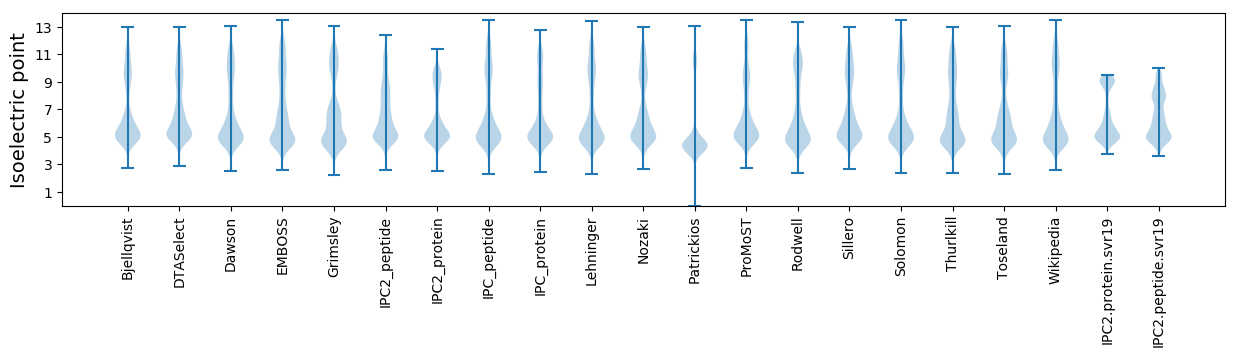
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M5R2U3|A0A1M5R2U3_9ACTN YD repeat-containing protein OS=Jatrophihabitans endophyticus OX=1206085 GN=SAMN05443575_3423 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
Molecular weight: 4.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1383085 |
29 |
7281 |
330.8 |
35.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.472 ± 0.064 | 0.727 ± 0.009 |
6.523 ± 0.037 | 4.798 ± 0.042 |
2.693 ± 0.026 | 9.438 ± 0.04 |
2.165 ± 0.019 | 3.076 ± 0.027 |
1.697 ± 0.029 | 10.031 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.517 ± 0.014 | 1.717 ± 0.02 |
5.7 ± 0.029 | 2.634 ± 0.022 |
8.22 ± 0.051 | 5.117 ± 0.041 |
6.473 ± 0.048 | 9.652 ± 0.037 |
1.379 ± 0.014 | 1.971 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
