
Nocardioides sp. HDW12B
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides; unclassified Nocardioides
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
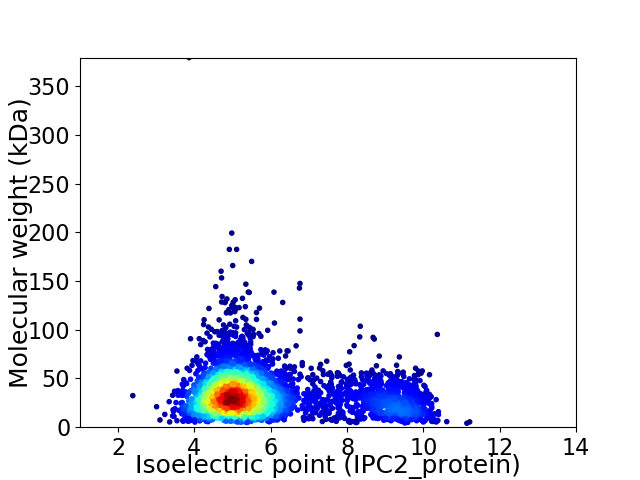
Virtual 2D-PAGE plot for 3834 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G7XQD2|A0A6G7XQD2_9ACTN 5-oxoprolinase subunit PxpB OS=Nocardioides sp. HDW12B OX=2714939 GN=pxpB PE=4 SV=1
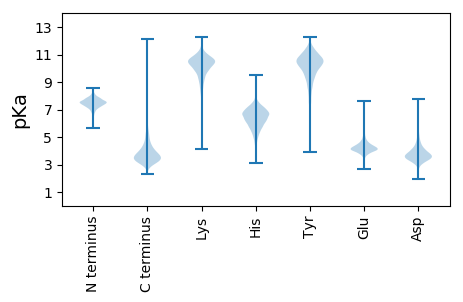
MM1 pKa = 7.75RR2 pKa = 11.84RR3 pKa = 11.84PKK5 pKa = 10.53SLLFAIPLTLALALTACSAEE25 pKa = 4.21SDD27 pKa = 3.82SSGGDD32 pKa = 3.21SSSEE36 pKa = 3.94SSSSSEE42 pKa = 3.95GGEE45 pKa = 4.28SAGTAGGTFSVSSSEE60 pKa = 4.29PEE62 pKa = 3.75AFAPTSNCYY71 pKa = 10.55SSDD74 pKa = 3.19CSQIINAVFTGLLKK88 pKa = 10.4VDD90 pKa = 4.63PEE92 pKa = 4.34TSEE95 pKa = 4.49PIYY98 pKa = 10.58TGAEE102 pKa = 4.02SFEE105 pKa = 4.62SEE107 pKa = 5.46DD108 pKa = 3.67GQNWTIKK115 pKa = 10.59LKK117 pKa = 11.04DD118 pKa = 3.23GFTFHH123 pKa = 7.05NGEE126 pKa = 4.08PVDD129 pKa = 3.62AEE131 pKa = 4.49SYY133 pKa = 9.16VRR135 pKa = 11.84AWNYY139 pKa = 9.38AANGANATQLGFFFSKK155 pKa = 11.01VEE157 pKa = 4.51GYY159 pKa = 11.04DD160 pKa = 3.58DD161 pKa = 4.5LQGEE165 pKa = 4.28NPKK168 pKa = 10.54AEE170 pKa = 4.28EE171 pKa = 4.03MSGLTVVDD179 pKa = 4.36DD180 pKa = 3.99LTFEE184 pKa = 4.39VALSEE189 pKa = 4.36PFSQFPLTMSYY200 pKa = 9.66TPAAAPMAQEE210 pKa = 4.52CMDD213 pKa = 5.56DD214 pKa = 3.68IKK216 pKa = 11.4ACNEE220 pKa = 3.73QPIGNGPYY228 pKa = 10.03QFAEE232 pKa = 4.22KK233 pKa = 9.59WNHH236 pKa = 4.8NQSITLEE243 pKa = 4.18KK244 pKa = 10.08YY245 pKa = 10.38ADD247 pKa = 3.73YY248 pKa = 11.11SDD250 pKa = 4.85EE251 pKa = 4.21EE252 pKa = 4.56TAGLADD258 pKa = 3.38GMEE261 pKa = 4.2FKK263 pKa = 10.38IYY265 pKa = 10.87ADD267 pKa = 3.79LSTAFRR273 pKa = 11.84DD274 pKa = 4.27FQGGNLDD281 pKa = 3.82IVIPDD286 pKa = 4.22PSQIPQARR294 pKa = 11.84AQYY297 pKa = 10.76ADD299 pKa = 3.52SLLEE303 pKa = 3.87VDD305 pKa = 4.58SGSFAYY311 pKa = 10.31FGFPFYY317 pKa = 10.94EE318 pKa = 4.58PDD320 pKa = 3.37FQKK323 pKa = 11.16PEE325 pKa = 3.53IRR327 pKa = 11.84QALSLAIDD335 pKa = 3.66RR336 pKa = 11.84QSIIDD341 pKa = 3.69NVLQGRR347 pKa = 11.84FIPATDD353 pKa = 3.81VISPFVPGSRR363 pKa = 11.84DD364 pKa = 3.55DD365 pKa = 3.9ACEE368 pKa = 3.77FCVYY372 pKa = 10.8DD373 pKa = 3.85PDD375 pKa = 4.01QAKK378 pKa = 10.01EE379 pKa = 3.92LWDD382 pKa = 3.68GAGGVEE388 pKa = 4.38GNEE391 pKa = 3.39ISIWFNNDD399 pKa = 2.37GGHH402 pKa = 5.05EE403 pKa = 4.04QWVKK407 pKa = 11.29AMAQGWQQDD416 pKa = 3.66LGVDD420 pKa = 4.32FTFKK424 pKa = 10.56SQPFTPYY431 pKa = 10.55LKK433 pKa = 10.84ALGTQSEE440 pKa = 4.41VDD442 pKa = 3.23GPYY445 pKa = 10.62RR446 pKa = 11.84LGWLPDD452 pKa = 3.49YY453 pKa = 9.91PSPQNYY459 pKa = 9.92LDD461 pKa = 3.86PLYY464 pKa = 11.32GEE466 pKa = 5.05GSSNYY471 pKa = 9.41GKK473 pKa = 9.81WGEE476 pKa = 4.11GDD478 pKa = 3.5SAADD482 pKa = 3.36QQEE485 pKa = 3.89FLDD488 pKa = 4.72LVNEE492 pKa = 4.3GDD494 pKa = 3.45AAATVEE500 pKa = 4.31EE501 pKa = 5.54GIPSYY506 pKa = 10.55QAAADD511 pKa = 3.78VVLDD515 pKa = 3.74NMVVIPLWFGQTFYY529 pKa = 11.9VMSEE533 pKa = 3.59NVEE536 pKa = 4.09NVKK539 pKa = 10.62YY540 pKa = 10.98SPTDD544 pKa = 3.17QLLLDD549 pKa = 5.27EE550 pKa = 5.26ISVTSS555 pKa = 3.62
MM1 pKa = 7.75RR2 pKa = 11.84RR3 pKa = 11.84PKK5 pKa = 10.53SLLFAIPLTLALALTACSAEE25 pKa = 4.21SDD27 pKa = 3.82SSGGDD32 pKa = 3.21SSSEE36 pKa = 3.94SSSSSEE42 pKa = 3.95GGEE45 pKa = 4.28SAGTAGGTFSVSSSEE60 pKa = 4.29PEE62 pKa = 3.75AFAPTSNCYY71 pKa = 10.55SSDD74 pKa = 3.19CSQIINAVFTGLLKK88 pKa = 10.4VDD90 pKa = 4.63PEE92 pKa = 4.34TSEE95 pKa = 4.49PIYY98 pKa = 10.58TGAEE102 pKa = 4.02SFEE105 pKa = 4.62SEE107 pKa = 5.46DD108 pKa = 3.67GQNWTIKK115 pKa = 10.59LKK117 pKa = 11.04DD118 pKa = 3.23GFTFHH123 pKa = 7.05NGEE126 pKa = 4.08PVDD129 pKa = 3.62AEE131 pKa = 4.49SYY133 pKa = 9.16VRR135 pKa = 11.84AWNYY139 pKa = 9.38AANGANATQLGFFFSKK155 pKa = 11.01VEE157 pKa = 4.51GYY159 pKa = 11.04DD160 pKa = 3.58DD161 pKa = 4.5LQGEE165 pKa = 4.28NPKK168 pKa = 10.54AEE170 pKa = 4.28EE171 pKa = 4.03MSGLTVVDD179 pKa = 4.36DD180 pKa = 3.99LTFEE184 pKa = 4.39VALSEE189 pKa = 4.36PFSQFPLTMSYY200 pKa = 9.66TPAAAPMAQEE210 pKa = 4.52CMDD213 pKa = 5.56DD214 pKa = 3.68IKK216 pKa = 11.4ACNEE220 pKa = 3.73QPIGNGPYY228 pKa = 10.03QFAEE232 pKa = 4.22KK233 pKa = 9.59WNHH236 pKa = 4.8NQSITLEE243 pKa = 4.18KK244 pKa = 10.08YY245 pKa = 10.38ADD247 pKa = 3.73YY248 pKa = 11.11SDD250 pKa = 4.85EE251 pKa = 4.21EE252 pKa = 4.56TAGLADD258 pKa = 3.38GMEE261 pKa = 4.2FKK263 pKa = 10.38IYY265 pKa = 10.87ADD267 pKa = 3.79LSTAFRR273 pKa = 11.84DD274 pKa = 4.27FQGGNLDD281 pKa = 3.82IVIPDD286 pKa = 4.22PSQIPQARR294 pKa = 11.84AQYY297 pKa = 10.76ADD299 pKa = 3.52SLLEE303 pKa = 3.87VDD305 pKa = 4.58SGSFAYY311 pKa = 10.31FGFPFYY317 pKa = 10.94EE318 pKa = 4.58PDD320 pKa = 3.37FQKK323 pKa = 11.16PEE325 pKa = 3.53IRR327 pKa = 11.84QALSLAIDD335 pKa = 3.66RR336 pKa = 11.84QSIIDD341 pKa = 3.69NVLQGRR347 pKa = 11.84FIPATDD353 pKa = 3.81VISPFVPGSRR363 pKa = 11.84DD364 pKa = 3.55DD365 pKa = 3.9ACEE368 pKa = 3.77FCVYY372 pKa = 10.8DD373 pKa = 3.85PDD375 pKa = 4.01QAKK378 pKa = 10.01EE379 pKa = 3.92LWDD382 pKa = 3.68GAGGVEE388 pKa = 4.38GNEE391 pKa = 3.39ISIWFNNDD399 pKa = 2.37GGHH402 pKa = 5.05EE403 pKa = 4.04QWVKK407 pKa = 11.29AMAQGWQQDD416 pKa = 3.66LGVDD420 pKa = 4.32FTFKK424 pKa = 10.56SQPFTPYY431 pKa = 10.55LKK433 pKa = 10.84ALGTQSEE440 pKa = 4.41VDD442 pKa = 3.23GPYY445 pKa = 10.62RR446 pKa = 11.84LGWLPDD452 pKa = 3.49YY453 pKa = 9.91PSPQNYY459 pKa = 9.92LDD461 pKa = 3.86PLYY464 pKa = 11.32GEE466 pKa = 5.05GSSNYY471 pKa = 9.41GKK473 pKa = 9.81WGEE476 pKa = 4.11GDD478 pKa = 3.5SAADD482 pKa = 3.36QQEE485 pKa = 3.89FLDD488 pKa = 4.72LVNEE492 pKa = 4.3GDD494 pKa = 3.45AAATVEE500 pKa = 4.31EE501 pKa = 5.54GIPSYY506 pKa = 10.55QAAADD511 pKa = 3.78VVLDD515 pKa = 3.74NMVVIPLWFGQTFYY529 pKa = 11.9VMSEE533 pKa = 3.59NVEE536 pKa = 4.09NVKK539 pKa = 10.62YY540 pKa = 10.98SPTDD544 pKa = 3.17QLLLDD549 pKa = 5.27EE550 pKa = 5.26ISVTSS555 pKa = 3.62
Molecular weight: 60.5 kDa
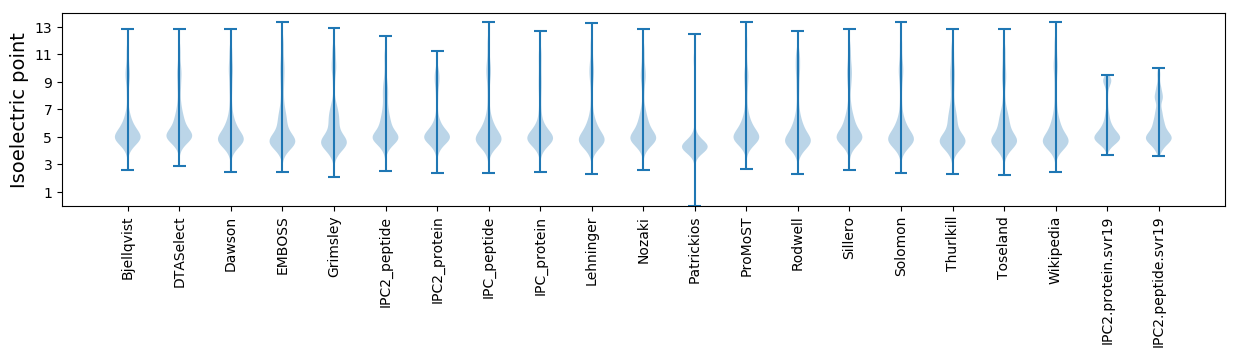
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G7XKU7|A0A6G7XKU7_9ACTN GNAT family N-acetyltransferase OS=Nocardioides sp. HDW12B OX=2714939 GN=G7072_01835 PE=4 SV=1
MM1 pKa = 7.28GSVVKK6 pKa = 10.49KK7 pKa = 9.45RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.48RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.12TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84LGKK33 pKa = 10.04
MM1 pKa = 7.28GSVVKK6 pKa = 10.49KK7 pKa = 9.45RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.48RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.12TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84LGKK33 pKa = 10.04
Molecular weight: 4.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1259540 |
33 |
3771 |
328.5 |
35.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.926 ± 0.061 | 0.723 ± 0.01 |
6.697 ± 0.032 | 5.996 ± 0.033 |
2.711 ± 0.021 | 9.419 ± 0.036 |
2.166 ± 0.019 | 2.915 ± 0.033 |
1.797 ± 0.034 | 10.304 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.755 ± 0.017 | 1.634 ± 0.024 |
5.748 ± 0.028 | 2.669 ± 0.019 |
7.956 ± 0.05 | 5.35 ± 0.029 |
6.151 ± 0.03 | 9.735 ± 0.041 |
1.485 ± 0.015 | 1.861 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
