
Culex tritaeniorhynchus rhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Merhavirus; Tritaeniorhynchus merhavirus
Average proteome isoelectric point is 7.32
Get precalculated fractions of proteins
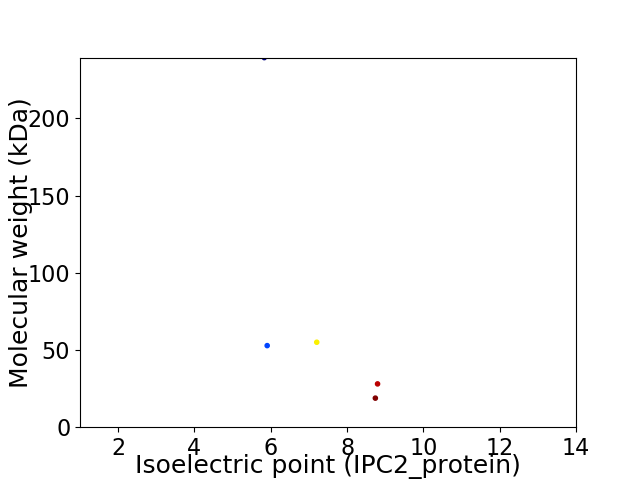
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions



Protein with the lowest isoelectric point:
>tr|F7J0V0|F7J0V0_9RHAB Phosphoprotein OS=Culex tritaeniorhynchus rhabdovirus OX=936308 GN=P PE=4 SV=2
MM1 pKa = 7.74SDD3 pKa = 3.51PVCRR7 pKa = 11.84HH8 pKa = 5.83GNLEE12 pKa = 4.44GYY14 pKa = 8.38QVPSISLASTIKK26 pKa = 9.7ATYY29 pKa = 9.19PSEE32 pKa = 4.69AFAKK36 pKa = 9.35QEE38 pKa = 3.93KK39 pKa = 9.89PIVRR43 pKa = 11.84GPAYY47 pKa = 9.64TGDD50 pKa = 3.59LAGFDD55 pKa = 4.75AYY57 pKa = 8.48MTSQLSQTTVTSLPIDD73 pKa = 3.52YY74 pKa = 10.34AICYY78 pKa = 8.02LHH80 pKa = 7.16AVACVTGVKK89 pKa = 10.37LNEE92 pKa = 3.96PWSSFGVNLTDD103 pKa = 4.6AEE105 pKa = 4.43NNVYY109 pKa = 9.98IASLYY114 pKa = 10.27KK115 pKa = 10.1FQAVGDD121 pKa = 4.14ALDD124 pKa = 3.91LSGGAVQGDD133 pKa = 3.57RR134 pKa = 11.84LYY136 pKa = 11.66NLFMLFAPCRR146 pKa = 11.84LHH148 pKa = 7.17SGLRR152 pKa = 11.84PEE154 pKa = 4.26YY155 pKa = 10.21RR156 pKa = 11.84PLLAEE161 pKa = 4.3RR162 pKa = 11.84YY163 pKa = 6.1KK164 pKa = 10.89TVMGDD169 pKa = 3.12KK170 pKa = 10.61KK171 pKa = 10.88KK172 pKa = 10.75GLEE175 pKa = 3.98PSALAIKK182 pKa = 10.13CAGWDD187 pKa = 3.3QHH189 pKa = 5.95PAYY192 pKa = 9.99AALAAAYY199 pKa = 10.76DD200 pKa = 3.54MFLFKK205 pKa = 10.66TEE207 pKa = 3.53NHH209 pKa = 6.58EE210 pKa = 4.13YY211 pKa = 10.46AKK213 pKa = 10.64VRR215 pKa = 11.84IGTTPMRR222 pKa = 11.84FRR224 pKa = 11.84DD225 pKa = 3.89CTMLGVLEE233 pKa = 4.22VMRR236 pKa = 11.84NAMKK240 pKa = 10.51VEE242 pKa = 4.05SMIDD246 pKa = 3.06ATYY249 pKa = 9.56WIWNGRR255 pKa = 11.84LADD258 pKa = 4.14EE259 pKa = 4.55FEE261 pKa = 4.85KK262 pKa = 10.24IYY264 pKa = 11.05KK265 pKa = 10.05EE266 pKa = 4.52GEE268 pKa = 4.11EE269 pKa = 4.02LDD271 pKa = 4.12KK272 pKa = 11.41ADD274 pKa = 5.07SYY276 pKa = 10.18TPYY279 pKa = 10.76YY280 pKa = 10.97AGFKK284 pKa = 10.09LGNRR288 pKa = 11.84SPYY291 pKa = 9.36SATMSPQLHH300 pKa = 5.92YY301 pKa = 10.15FVHH304 pKa = 6.99TIGCLANSKK313 pKa = 10.42RR314 pKa = 11.84SINARR319 pKa = 11.84AITEE323 pKa = 4.01VGIEE327 pKa = 4.08DD328 pKa = 3.91ARR330 pKa = 11.84DD331 pKa = 3.03NAIIVGYY338 pKa = 10.51VMAGRR343 pKa = 11.84RR344 pKa = 11.84RR345 pKa = 11.84HH346 pKa = 5.19RR347 pKa = 11.84AQYY350 pKa = 9.35FRR352 pKa = 11.84PSEE355 pKa = 4.26AEE357 pKa = 3.62LWKK360 pKa = 10.83APEE363 pKa = 3.92PRR365 pKa = 11.84PRR367 pKa = 11.84ADD369 pKa = 4.03CLPGANEE376 pKa = 3.93AVRR379 pKa = 11.84DD380 pKa = 3.82RR381 pKa = 11.84GQEE384 pKa = 3.91DD385 pKa = 4.13EE386 pKa = 4.44EE387 pKa = 5.49SSVSEE392 pKa = 4.1VSSVCSRR399 pKa = 11.84IARR402 pKa = 11.84DD403 pKa = 3.44FDD405 pKa = 3.86GEE407 pKa = 4.47EE408 pKa = 3.81IPEE411 pKa = 4.37PLSSRR416 pKa = 11.84ASDD419 pKa = 3.64WYY421 pKa = 10.89QYY423 pKa = 9.03MASQHH428 pKa = 6.43FIMPRR433 pKa = 11.84QMEE436 pKa = 3.99IAACNILAALDD447 pKa = 3.49KK448 pKa = 10.49TRR450 pKa = 11.84PDD452 pKa = 3.73TIGDD456 pKa = 4.06HH457 pKa = 6.35LKK459 pKa = 8.79MWAKK463 pKa = 10.67AKK465 pKa = 10.49LRR467 pKa = 11.84TLQQTT472 pKa = 3.54
MM1 pKa = 7.74SDD3 pKa = 3.51PVCRR7 pKa = 11.84HH8 pKa = 5.83GNLEE12 pKa = 4.44GYY14 pKa = 8.38QVPSISLASTIKK26 pKa = 9.7ATYY29 pKa = 9.19PSEE32 pKa = 4.69AFAKK36 pKa = 9.35QEE38 pKa = 3.93KK39 pKa = 9.89PIVRR43 pKa = 11.84GPAYY47 pKa = 9.64TGDD50 pKa = 3.59LAGFDD55 pKa = 4.75AYY57 pKa = 8.48MTSQLSQTTVTSLPIDD73 pKa = 3.52YY74 pKa = 10.34AICYY78 pKa = 8.02LHH80 pKa = 7.16AVACVTGVKK89 pKa = 10.37LNEE92 pKa = 3.96PWSSFGVNLTDD103 pKa = 4.6AEE105 pKa = 4.43NNVYY109 pKa = 9.98IASLYY114 pKa = 10.27KK115 pKa = 10.1FQAVGDD121 pKa = 4.14ALDD124 pKa = 3.91LSGGAVQGDD133 pKa = 3.57RR134 pKa = 11.84LYY136 pKa = 11.66NLFMLFAPCRR146 pKa = 11.84LHH148 pKa = 7.17SGLRR152 pKa = 11.84PEE154 pKa = 4.26YY155 pKa = 10.21RR156 pKa = 11.84PLLAEE161 pKa = 4.3RR162 pKa = 11.84YY163 pKa = 6.1KK164 pKa = 10.89TVMGDD169 pKa = 3.12KK170 pKa = 10.61KK171 pKa = 10.88KK172 pKa = 10.75GLEE175 pKa = 3.98PSALAIKK182 pKa = 10.13CAGWDD187 pKa = 3.3QHH189 pKa = 5.95PAYY192 pKa = 9.99AALAAAYY199 pKa = 10.76DD200 pKa = 3.54MFLFKK205 pKa = 10.66TEE207 pKa = 3.53NHH209 pKa = 6.58EE210 pKa = 4.13YY211 pKa = 10.46AKK213 pKa = 10.64VRR215 pKa = 11.84IGTTPMRR222 pKa = 11.84FRR224 pKa = 11.84DD225 pKa = 3.89CTMLGVLEE233 pKa = 4.22VMRR236 pKa = 11.84NAMKK240 pKa = 10.51VEE242 pKa = 4.05SMIDD246 pKa = 3.06ATYY249 pKa = 9.56WIWNGRR255 pKa = 11.84LADD258 pKa = 4.14EE259 pKa = 4.55FEE261 pKa = 4.85KK262 pKa = 10.24IYY264 pKa = 11.05KK265 pKa = 10.05EE266 pKa = 4.52GEE268 pKa = 4.11EE269 pKa = 4.02LDD271 pKa = 4.12KK272 pKa = 11.41ADD274 pKa = 5.07SYY276 pKa = 10.18TPYY279 pKa = 10.76YY280 pKa = 10.97AGFKK284 pKa = 10.09LGNRR288 pKa = 11.84SPYY291 pKa = 9.36SATMSPQLHH300 pKa = 5.92YY301 pKa = 10.15FVHH304 pKa = 6.99TIGCLANSKK313 pKa = 10.42RR314 pKa = 11.84SINARR319 pKa = 11.84AITEE323 pKa = 4.01VGIEE327 pKa = 4.08DD328 pKa = 3.91ARR330 pKa = 11.84DD331 pKa = 3.03NAIIVGYY338 pKa = 10.51VMAGRR343 pKa = 11.84RR344 pKa = 11.84RR345 pKa = 11.84HH346 pKa = 5.19RR347 pKa = 11.84AQYY350 pKa = 9.35FRR352 pKa = 11.84PSEE355 pKa = 4.26AEE357 pKa = 3.62LWKK360 pKa = 10.83APEE363 pKa = 3.92PRR365 pKa = 11.84PRR367 pKa = 11.84ADD369 pKa = 4.03CLPGANEE376 pKa = 3.93AVRR379 pKa = 11.84DD380 pKa = 3.82RR381 pKa = 11.84GQEE384 pKa = 3.91DD385 pKa = 4.13EE386 pKa = 4.44EE387 pKa = 5.49SSVSEE392 pKa = 4.1VSSVCSRR399 pKa = 11.84IARR402 pKa = 11.84DD403 pKa = 3.44FDD405 pKa = 3.86GEE407 pKa = 4.47EE408 pKa = 3.81IPEE411 pKa = 4.37PLSSRR416 pKa = 11.84ASDD419 pKa = 3.64WYY421 pKa = 10.89QYY423 pKa = 9.03MASQHH428 pKa = 6.43FIMPRR433 pKa = 11.84QMEE436 pKa = 3.99IAACNILAALDD447 pKa = 3.49KK448 pKa = 10.49TRR450 pKa = 11.84PDD452 pKa = 3.73TIGDD456 pKa = 4.06HH457 pKa = 6.35LKK459 pKa = 8.79MWAKK463 pKa = 10.67AKK465 pKa = 10.49LRR467 pKa = 11.84TLQQTT472 pKa = 3.54
Molecular weight: 52.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F7J0V2|F7J0V2_9RHAB Glycoprotein OS=Culex tritaeniorhynchus rhabdovirus OX=936308 GN=G PE=4 SV=1
MM1 pKa = 7.1STSQVGVKK9 pKa = 9.66SFKK12 pKa = 10.51ILFDD16 pKa = 3.96LQLSSDD22 pKa = 3.48TRR24 pKa = 11.84FNNCLDD30 pKa = 4.17LACGISEE37 pKa = 4.18VRR39 pKa = 11.84DD40 pKa = 3.62AMRR43 pKa = 11.84GTPTEE48 pKa = 3.89VSFLLGVLGLGLYY61 pKa = 9.89GSKK64 pKa = 10.66AEE66 pKa = 4.26KK67 pKa = 10.15LAAGGSWSKK76 pKa = 10.53RR77 pKa = 11.84VEE79 pKa = 4.76DD80 pKa = 3.49IVKK83 pKa = 9.46TKK85 pKa = 9.85TAVALLKK92 pKa = 10.93SPIARR97 pKa = 11.84SFRR100 pKa = 11.84YY101 pKa = 9.7EE102 pKa = 3.44NSLTLRR108 pKa = 11.84GARR111 pKa = 11.84TYY113 pKa = 11.05IRR115 pKa = 11.84MSVQISEE122 pKa = 4.31TTLDD126 pKa = 3.1GRR128 pKa = 11.84RR129 pKa = 11.84YY130 pKa = 9.86RR131 pKa = 11.84EE132 pKa = 3.87VFGLMDD138 pKa = 4.35KK139 pKa = 11.12GKK141 pKa = 7.72PTPKK145 pKa = 10.64GPIKK149 pKa = 10.35EE150 pKa = 4.15VLAVFGVTLEE160 pKa = 4.37SVGGLDD166 pKa = 3.98EE167 pKa = 5.95LSMISLL173 pKa = 4.34
MM1 pKa = 7.1STSQVGVKK9 pKa = 9.66SFKK12 pKa = 10.51ILFDD16 pKa = 3.96LQLSSDD22 pKa = 3.48TRR24 pKa = 11.84FNNCLDD30 pKa = 4.17LACGISEE37 pKa = 4.18VRR39 pKa = 11.84DD40 pKa = 3.62AMRR43 pKa = 11.84GTPTEE48 pKa = 3.89VSFLLGVLGLGLYY61 pKa = 9.89GSKK64 pKa = 10.66AEE66 pKa = 4.26KK67 pKa = 10.15LAAGGSWSKK76 pKa = 10.53RR77 pKa = 11.84VEE79 pKa = 4.76DD80 pKa = 3.49IVKK83 pKa = 9.46TKK85 pKa = 9.85TAVALLKK92 pKa = 10.93SPIARR97 pKa = 11.84SFRR100 pKa = 11.84YY101 pKa = 9.7EE102 pKa = 3.44NSLTLRR108 pKa = 11.84GARR111 pKa = 11.84TYY113 pKa = 11.05IRR115 pKa = 11.84MSVQISEE122 pKa = 4.31TTLDD126 pKa = 3.1GRR128 pKa = 11.84RR129 pKa = 11.84YY130 pKa = 9.86RR131 pKa = 11.84EE132 pKa = 3.87VFGLMDD138 pKa = 4.35KK139 pKa = 11.12GKK141 pKa = 7.72PTPKK145 pKa = 10.64GPIKK149 pKa = 10.35EE150 pKa = 4.15VLAVFGVTLEE160 pKa = 4.37SVGGLDD166 pKa = 3.98EE167 pKa = 5.95LSMISLL173 pKa = 4.34
Molecular weight: 18.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3535 |
173 |
2123 |
707.0 |
78.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.496 ± 0.995 | 1.754 ± 0.221 |
5.516 ± 0.612 | 5.941 ± 0.21 |
3.876 ± 0.534 | 7.666 ± 0.843 |
2.405 ± 0.367 | 5.969 ± 0.554 |
4.668 ± 0.432 | 9.901 ± 0.625 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.207 ± 0.28 | 3.338 ± 0.512 |
5.488 ± 0.691 | 2.744 ± 0.311 |
6.478 ± 0.201 | 7.61 ± 1.104 |
5.827 ± 0.301 | 5.912 ± 0.402 |
2.008 ± 0.269 | 3.197 ± 0.479 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
