
Bacillus sp. OK048
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; unclassified Bacillus (in: Bacteria)
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
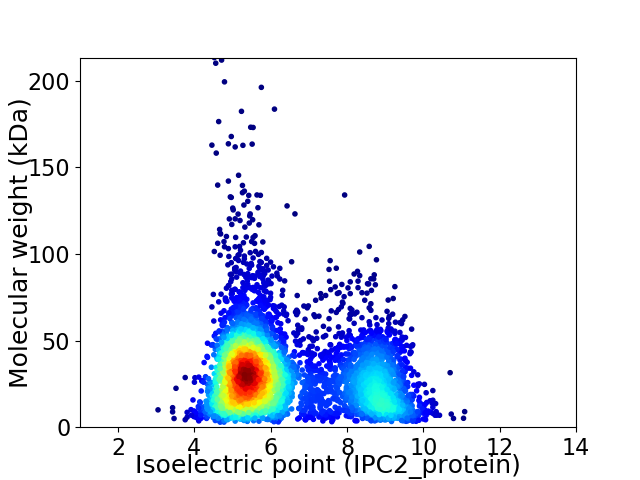
Virtual 2D-PAGE plot for 5056 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H0BN61|A0A1H0BN61_9BACI Acetoacetyl-CoA synthetase OS=Bacillus sp. OK048 OX=1882761 GN=SAMN05443253_11293 PE=3 SV=1
MM1 pKa = 7.5SLEE4 pKa = 3.92WFDD7 pKa = 4.15RR8 pKa = 11.84VCSEE12 pKa = 4.49LQDD15 pKa = 3.73HH16 pKa = 6.83LEE18 pKa = 4.58SICEE22 pKa = 4.06EE23 pKa = 4.28YY24 pKa = 11.0NQVGQMSIEE33 pKa = 4.15RR34 pKa = 11.84ASKK37 pKa = 9.74HH38 pKa = 4.75PRR40 pKa = 11.84IEE42 pKa = 4.2FFVEE46 pKa = 4.12TEE48 pKa = 4.03DD49 pKa = 4.56VQTEE53 pKa = 4.42DD54 pKa = 2.75IDD56 pKa = 4.45RR57 pKa = 11.84DD58 pKa = 4.05YY59 pKa = 11.54FCTLFFDD66 pKa = 4.45PGNEE70 pKa = 3.63EE71 pKa = 4.51FYY73 pKa = 10.66IDD75 pKa = 4.07TFDD78 pKa = 3.5VDD80 pKa = 3.74SGHH83 pKa = 5.37TAKK86 pKa = 10.56IILSDD91 pKa = 3.23IEE93 pKa = 4.26EE94 pKa = 4.91IIDD97 pKa = 3.59EE98 pKa = 4.34VHH100 pKa = 6.94ISLHH104 pKa = 7.34DD105 pKa = 3.71YY106 pKa = 10.9MNDD109 pKa = 3.29DD110 pKa = 4.43DD111 pKa = 6.19HH112 pKa = 6.46YY113 pKa = 9.45TDD115 pKa = 4.69DD116 pKa = 4.79GVYY119 pKa = 8.61LTSNEE124 pKa = 4.09VYY126 pKa = 10.2EE127 pKa = 5.73DD128 pKa = 3.8SDD130 pKa = 3.47DD131 pKa = 4.1CYY133 pKa = 11.82EE134 pKa = 4.07MVEE137 pKa = 4.02VDD139 pKa = 4.32DD140 pKa = 6.33DD141 pKa = 4.12GVEE144 pKa = 4.31YY145 pKa = 10.87YY146 pKa = 10.15QAEE149 pKa = 4.39QVNSDD154 pKa = 4.77DD155 pKa = 3.48IFEE158 pKa = 5.14EE159 pKa = 5.19IDD161 pKa = 3.58VDD163 pKa = 3.77WNTPEE168 pKa = 3.74VTAFKK173 pKa = 10.4HH174 pKa = 5.13GNEE177 pKa = 4.16VEE179 pKa = 4.1VTYY182 pKa = 10.92QFGVVSEE189 pKa = 4.42TGDD192 pKa = 3.46GVLKK196 pKa = 10.46RR197 pKa = 11.84INRR200 pKa = 11.84IWTTNDD206 pKa = 2.63EE207 pKa = 4.75LIRR210 pKa = 11.84DD211 pKa = 3.73EE212 pKa = 4.36SHH214 pKa = 7.34FIFSKK219 pKa = 10.93EE220 pKa = 3.83EE221 pKa = 3.63ASTIIAMIASHH232 pKa = 6.76MDD234 pKa = 3.12QLSEE238 pKa = 4.74FNFDD242 pKa = 5.1DD243 pKa = 3.93MPP245 pKa = 6.42
MM1 pKa = 7.5SLEE4 pKa = 3.92WFDD7 pKa = 4.15RR8 pKa = 11.84VCSEE12 pKa = 4.49LQDD15 pKa = 3.73HH16 pKa = 6.83LEE18 pKa = 4.58SICEE22 pKa = 4.06EE23 pKa = 4.28YY24 pKa = 11.0NQVGQMSIEE33 pKa = 4.15RR34 pKa = 11.84ASKK37 pKa = 9.74HH38 pKa = 4.75PRR40 pKa = 11.84IEE42 pKa = 4.2FFVEE46 pKa = 4.12TEE48 pKa = 4.03DD49 pKa = 4.56VQTEE53 pKa = 4.42DD54 pKa = 2.75IDD56 pKa = 4.45RR57 pKa = 11.84DD58 pKa = 4.05YY59 pKa = 11.54FCTLFFDD66 pKa = 4.45PGNEE70 pKa = 3.63EE71 pKa = 4.51FYY73 pKa = 10.66IDD75 pKa = 4.07TFDD78 pKa = 3.5VDD80 pKa = 3.74SGHH83 pKa = 5.37TAKK86 pKa = 10.56IILSDD91 pKa = 3.23IEE93 pKa = 4.26EE94 pKa = 4.91IIDD97 pKa = 3.59EE98 pKa = 4.34VHH100 pKa = 6.94ISLHH104 pKa = 7.34DD105 pKa = 3.71YY106 pKa = 10.9MNDD109 pKa = 3.29DD110 pKa = 4.43DD111 pKa = 6.19HH112 pKa = 6.46YY113 pKa = 9.45TDD115 pKa = 4.69DD116 pKa = 4.79GVYY119 pKa = 8.61LTSNEE124 pKa = 4.09VYY126 pKa = 10.2EE127 pKa = 5.73DD128 pKa = 3.8SDD130 pKa = 3.47DD131 pKa = 4.1CYY133 pKa = 11.82EE134 pKa = 4.07MVEE137 pKa = 4.02VDD139 pKa = 4.32DD140 pKa = 6.33DD141 pKa = 4.12GVEE144 pKa = 4.31YY145 pKa = 10.87YY146 pKa = 10.15QAEE149 pKa = 4.39QVNSDD154 pKa = 4.77DD155 pKa = 3.48IFEE158 pKa = 5.14EE159 pKa = 5.19IDD161 pKa = 3.58VDD163 pKa = 3.77WNTPEE168 pKa = 3.74VTAFKK173 pKa = 10.4HH174 pKa = 5.13GNEE177 pKa = 4.16VEE179 pKa = 4.1VTYY182 pKa = 10.92QFGVVSEE189 pKa = 4.42TGDD192 pKa = 3.46GVLKK196 pKa = 10.46RR197 pKa = 11.84INRR200 pKa = 11.84IWTTNDD206 pKa = 2.63EE207 pKa = 4.75LIRR210 pKa = 11.84DD211 pKa = 3.73EE212 pKa = 4.36SHH214 pKa = 7.34FIFSKK219 pKa = 10.93EE220 pKa = 3.83EE221 pKa = 3.63ASTIIAMIASHH232 pKa = 6.76MDD234 pKa = 3.12QLSEE238 pKa = 4.74FNFDD242 pKa = 5.1DD243 pKa = 3.93MPP245 pKa = 6.42
Molecular weight: 28.66 kDa
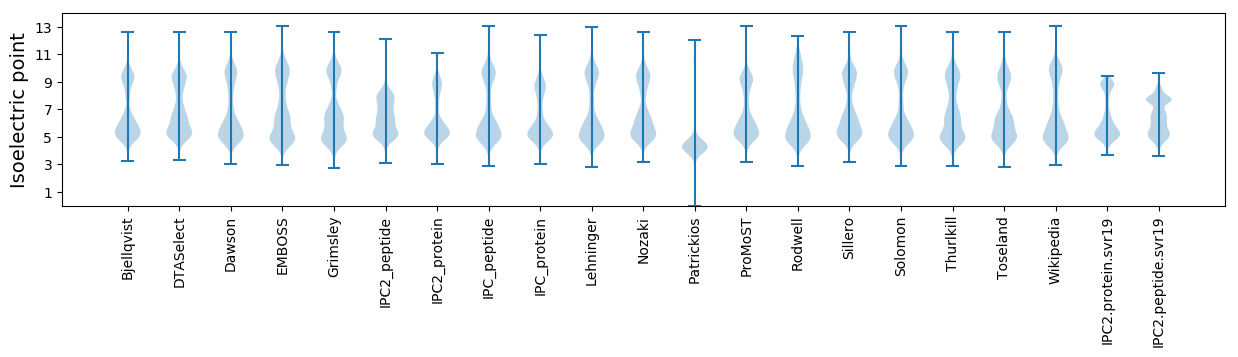
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H0CZM9|A0A1H0CZM9_9BACI EthD domain-containing protein OS=Bacillus sp. OK048 OX=1882761 GN=SAMN05443253_11558 PE=4 SV=1
MM1 pKa = 7.79PFYY4 pKa = 10.32PPNPGRR10 pKa = 11.84RR11 pKa = 11.84PFPPGNRR18 pKa = 11.84QRR20 pKa = 11.84FGPSNQMGRR29 pKa = 11.84RR30 pKa = 11.84PRR32 pKa = 11.84PFNVPNQNQQAPTSRR47 pKa = 11.84FQGLKK52 pKa = 10.98AMMGHH57 pKa = 6.48VGTVRR62 pKa = 11.84SGINTLRR69 pKa = 11.84QVGSFLKK76 pKa = 10.53FFKK79 pKa = 10.82
MM1 pKa = 7.79PFYY4 pKa = 10.32PPNPGRR10 pKa = 11.84RR11 pKa = 11.84PFPPGNRR18 pKa = 11.84QRR20 pKa = 11.84FGPSNQMGRR29 pKa = 11.84RR30 pKa = 11.84PRR32 pKa = 11.84PFNVPNQNQQAPTSRR47 pKa = 11.84FQGLKK52 pKa = 10.98AMMGHH57 pKa = 6.48VGTVRR62 pKa = 11.84SGINTLRR69 pKa = 11.84QVGSFLKK76 pKa = 10.53FFKK79 pKa = 10.82
Molecular weight: 8.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1448404 |
26 |
1961 |
286.5 |
32.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.731 ± 0.04 | 0.748 ± 0.011 |
4.961 ± 0.028 | 7.305 ± 0.048 |
4.718 ± 0.032 | 6.938 ± 0.042 |
2.038 ± 0.017 | 8.105 ± 0.039 |
7.12 ± 0.033 | 9.78 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.73 ± 0.017 | 4.724 ± 0.028 |
3.621 ± 0.021 | 3.604 ± 0.024 |
3.923 ± 0.028 | 6.021 ± 0.023 |
5.428 ± 0.027 | 6.942 ± 0.028 |
1.064 ± 0.014 | 3.502 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
