
Gammaproteobacteria bacterium LSUCC0057
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Porticoccaceae; unclassified Porticoccaceae; SAR92 clade
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
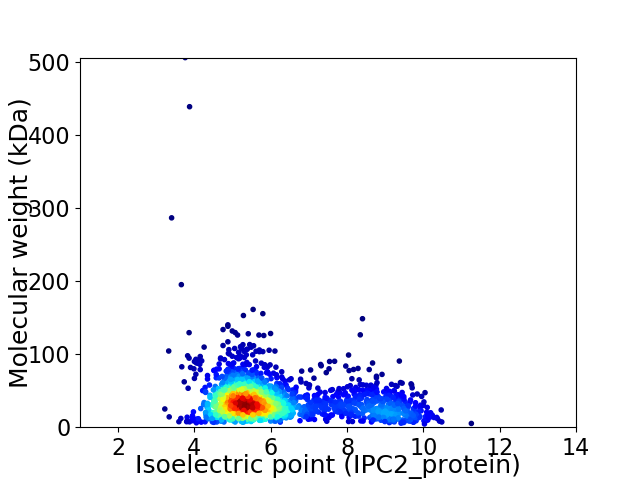
Virtual 2D-PAGE plot for 1957 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y8UFT2|A0A4Y8UFT2_9GAMM Cytochrome c domain-containing protein OS=Gammaproteobacteria bacterium LSUCC0057 OX=2559237 GN=E3W66_09575 PE=4 SV=1
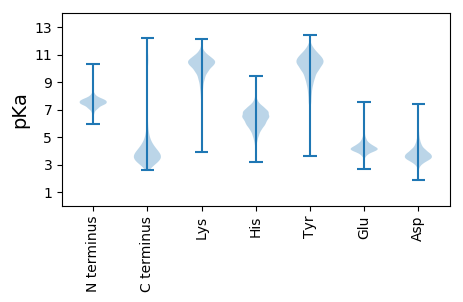
MM1 pKa = 7.79AIKK4 pKa = 10.56KK5 pKa = 9.73LVLVSAISGLLTACGGGDD23 pKa = 3.77VNISPSNVDD32 pKa = 3.12NSVDD36 pKa = 3.46NSTTTTGGSSSNGFCASYY54 pKa = 10.36TKK56 pKa = 10.48DD57 pKa = 3.08AQLYY61 pKa = 9.0EE62 pKa = 4.44GTLSGGNCVYY72 pKa = 10.39EE73 pKa = 4.44RR74 pKa = 11.84NFVDD78 pKa = 2.93ITNPLTVDD86 pKa = 3.21VTFPSIGSGKK96 pKa = 9.76HH97 pKa = 4.01VFEE100 pKa = 5.2GSLVVGEE107 pKa = 5.11AYY109 pKa = 10.46DD110 pKa = 4.52SIADD114 pKa = 3.83LTAAGITEE122 pKa = 4.59GGDD125 pKa = 3.17GATITIEE132 pKa = 4.03AGNTFVFKK140 pKa = 10.38TSEE143 pKa = 4.08DD144 pKa = 3.75YY145 pKa = 11.36AVINRR150 pKa = 11.84GSQIIAIGDD159 pKa = 3.67AEE161 pKa = 4.25NPITFTSYY169 pKa = 11.02SDD171 pKa = 4.51AISGTVAFDD180 pKa = 5.96DD181 pKa = 4.01VQQWGGMIINGFGVTNKK198 pKa = 9.91CAYY201 pKa = 9.11TGDD204 pKa = 3.41RR205 pKa = 11.84GAADD209 pKa = 3.78FAMVGGEE216 pKa = 4.21CNVAAEE222 pKa = 4.72GKK224 pKa = 9.89SGSAQTYY231 pKa = 9.49YY232 pKa = 11.16GGDD235 pKa = 3.44NDD237 pKa = 4.73ADD239 pKa = 3.88SSGHH243 pKa = 6.06LEE245 pKa = 4.06YY246 pKa = 10.53FIVKK250 pKa = 8.43HH251 pKa = 5.78TGAEE255 pKa = 4.04VAPGNEE261 pKa = 4.04LNGIAFGGVGSGTTVNYY278 pKa = 9.75LQVYY282 pKa = 7.32STYY285 pKa = 11.14DD286 pKa = 3.1DD287 pKa = 4.67GIEE290 pKa = 3.99MFGGSVDD297 pKa = 3.38VSNFVALYY305 pKa = 10.6VRR307 pKa = 11.84DD308 pKa = 4.08DD309 pKa = 3.82SFDD312 pKa = 3.24VDD314 pKa = 3.19EE315 pKa = 5.83GYY317 pKa = 11.24NGTLQNALIIQANDD331 pKa = 3.72DD332 pKa = 3.72GQQCIEE338 pKa = 3.9SDD340 pKa = 4.15GIGSYY345 pKa = 10.78DD346 pKa = 3.42SSEE349 pKa = 4.14TTRR352 pKa = 11.84NADD355 pKa = 4.7FIARR359 pKa = 11.84GLNSAATVNNMTCIVSGNQGGTHH382 pKa = 6.58GDD384 pKa = 3.49SVGVRR389 pKa = 11.84IRR391 pKa = 11.84EE392 pKa = 3.95GHH394 pKa = 7.24AITINDD400 pKa = 4.26SIITSALTNNDD411 pKa = 3.32GLDD414 pKa = 3.22EE415 pKa = 4.35CLRR418 pKa = 11.84IDD420 pKa = 4.98DD421 pKa = 4.79AEE423 pKa = 4.14TRR425 pKa = 11.84QAGFVTITNTLLACKK440 pKa = 10.25KK441 pKa = 9.27PFKK444 pKa = 10.77DD445 pKa = 3.07EE446 pKa = 3.82AVQIGSVGIEE456 pKa = 3.77AWFEE460 pKa = 3.75AADD463 pKa = 3.7GDD465 pKa = 4.31NVVEE469 pKa = 4.38RR470 pKa = 11.84TDD472 pKa = 3.4GAQTITEE479 pKa = 4.12LSGYY483 pKa = 10.14YY484 pKa = 8.81STGDD488 pKa = 3.19RR489 pKa = 11.84GAVTAADD496 pKa = 4.02DD497 pKa = 3.76WTEE500 pKa = 3.82GWTVGVDD507 pKa = 3.79ATWVTPP513 pKa = 3.85
MM1 pKa = 7.79AIKK4 pKa = 10.56KK5 pKa = 9.73LVLVSAISGLLTACGGGDD23 pKa = 3.77VNISPSNVDD32 pKa = 3.12NSVDD36 pKa = 3.46NSTTTTGGSSSNGFCASYY54 pKa = 10.36TKK56 pKa = 10.48DD57 pKa = 3.08AQLYY61 pKa = 9.0EE62 pKa = 4.44GTLSGGNCVYY72 pKa = 10.39EE73 pKa = 4.44RR74 pKa = 11.84NFVDD78 pKa = 2.93ITNPLTVDD86 pKa = 3.21VTFPSIGSGKK96 pKa = 9.76HH97 pKa = 4.01VFEE100 pKa = 5.2GSLVVGEE107 pKa = 5.11AYY109 pKa = 10.46DD110 pKa = 4.52SIADD114 pKa = 3.83LTAAGITEE122 pKa = 4.59GGDD125 pKa = 3.17GATITIEE132 pKa = 4.03AGNTFVFKK140 pKa = 10.38TSEE143 pKa = 4.08DD144 pKa = 3.75YY145 pKa = 11.36AVINRR150 pKa = 11.84GSQIIAIGDD159 pKa = 3.67AEE161 pKa = 4.25NPITFTSYY169 pKa = 11.02SDD171 pKa = 4.51AISGTVAFDD180 pKa = 5.96DD181 pKa = 4.01VQQWGGMIINGFGVTNKK198 pKa = 9.91CAYY201 pKa = 9.11TGDD204 pKa = 3.41RR205 pKa = 11.84GAADD209 pKa = 3.78FAMVGGEE216 pKa = 4.21CNVAAEE222 pKa = 4.72GKK224 pKa = 9.89SGSAQTYY231 pKa = 9.49YY232 pKa = 11.16GGDD235 pKa = 3.44NDD237 pKa = 4.73ADD239 pKa = 3.88SSGHH243 pKa = 6.06LEE245 pKa = 4.06YY246 pKa = 10.53FIVKK250 pKa = 8.43HH251 pKa = 5.78TGAEE255 pKa = 4.04VAPGNEE261 pKa = 4.04LNGIAFGGVGSGTTVNYY278 pKa = 9.75LQVYY282 pKa = 7.32STYY285 pKa = 11.14DD286 pKa = 3.1DD287 pKa = 4.67GIEE290 pKa = 3.99MFGGSVDD297 pKa = 3.38VSNFVALYY305 pKa = 10.6VRR307 pKa = 11.84DD308 pKa = 4.08DD309 pKa = 3.82SFDD312 pKa = 3.24VDD314 pKa = 3.19EE315 pKa = 5.83GYY317 pKa = 11.24NGTLQNALIIQANDD331 pKa = 3.72DD332 pKa = 3.72GQQCIEE338 pKa = 3.9SDD340 pKa = 4.15GIGSYY345 pKa = 10.78DD346 pKa = 3.42SSEE349 pKa = 4.14TTRR352 pKa = 11.84NADD355 pKa = 4.7FIARR359 pKa = 11.84GLNSAATVNNMTCIVSGNQGGTHH382 pKa = 6.58GDD384 pKa = 3.49SVGVRR389 pKa = 11.84IRR391 pKa = 11.84EE392 pKa = 3.95GHH394 pKa = 7.24AITINDD400 pKa = 4.26SIITSALTNNDD411 pKa = 3.32GLDD414 pKa = 3.22EE415 pKa = 4.35CLRR418 pKa = 11.84IDD420 pKa = 4.98DD421 pKa = 4.79AEE423 pKa = 4.14TRR425 pKa = 11.84QAGFVTITNTLLACKK440 pKa = 10.25KK441 pKa = 9.27PFKK444 pKa = 10.77DD445 pKa = 3.07EE446 pKa = 3.82AVQIGSVGIEE456 pKa = 3.77AWFEE460 pKa = 3.75AADD463 pKa = 3.7GDD465 pKa = 4.31NVVEE469 pKa = 4.38RR470 pKa = 11.84TDD472 pKa = 3.4GAQTITEE479 pKa = 4.12LSGYY483 pKa = 10.14YY484 pKa = 8.81STGDD488 pKa = 3.19RR489 pKa = 11.84GAVTAADD496 pKa = 4.02DD497 pKa = 3.76WTEE500 pKa = 3.82GWTVGVDD507 pKa = 3.79ATWVTPP513 pKa = 3.85
Molecular weight: 53.34 kDa
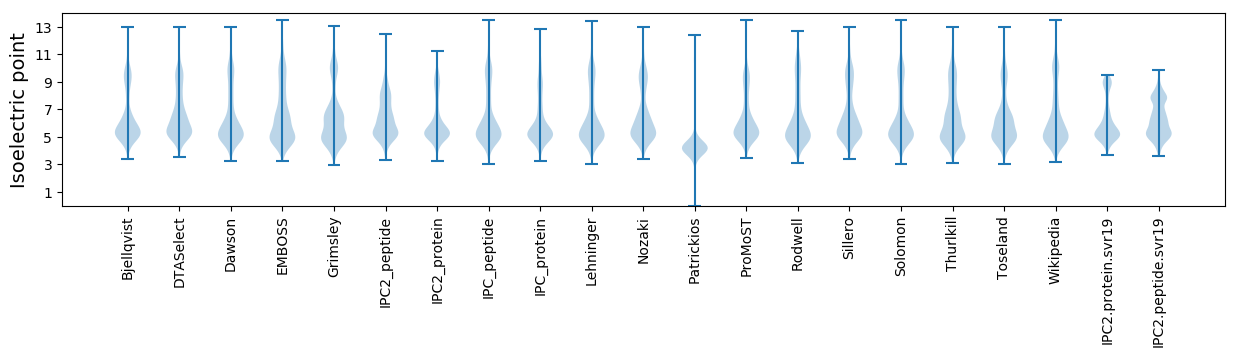
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y8UH41|A0A4Y8UH41_9GAMM Glucose 1-dehydrogenase OS=Gammaproteobacteria bacterium LSUCC0057 OX=2559237 GN=E3W66_05720 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.52RR12 pKa = 11.84KK13 pKa = 8.26RR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.16NGRR28 pKa = 11.84AVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.02GRR39 pKa = 11.84KK40 pKa = 8.77RR41 pKa = 11.84LAAA44 pKa = 4.42
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.52RR12 pKa = 11.84KK13 pKa = 8.26RR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.16NGRR28 pKa = 11.84AVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.02GRR39 pKa = 11.84KK40 pKa = 8.77RR41 pKa = 11.84LAAA44 pKa = 4.42
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
679377 |
38 |
5070 |
347.2 |
37.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.214 ± 0.063 | 1.078 ± 0.02 |
5.407 ± 0.051 | 5.588 ± 0.049 |
3.4 ± 0.031 | 7.792 ± 0.054 |
2.133 ± 0.03 | 4.967 ± 0.044 |
2.861 ± 0.042 | 11.28 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.11 ± 0.029 | 3.098 ± 0.035 |
4.513 ± 0.04 | 5.203 ± 0.053 |
6.226 ± 0.059 | 6.24 ± 0.059 |
4.796 ± 0.056 | 7.086 ± 0.043 |
1.348 ± 0.021 | 2.659 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
