
Hallerella succinigenes
Taxonomy: cellular organisms; Bacteria; FCB group; Fibrobacteres; Fibrobacteria; Fibrobacterales; Fibrobacteraceae; Hallerella
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
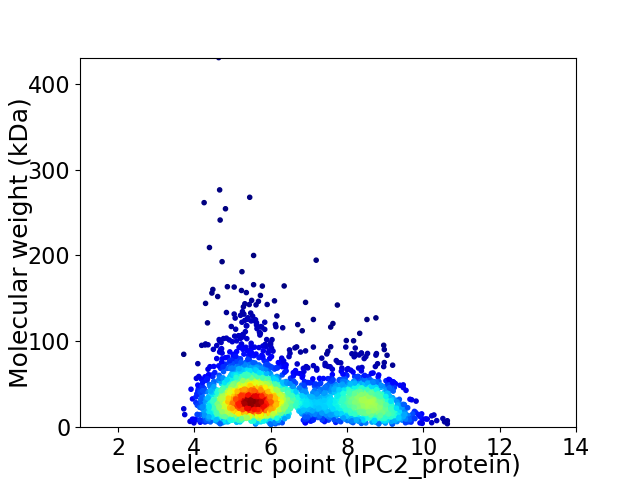
Virtual 2D-PAGE plot for 2694 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2M9A7Q5|A0A2M9A7Q5_9BACT Thioredoxin reductase (NADPH) OS=Hallerella succinigenes OX=1896222 GN=BGX16_1651 PE=4 SV=1
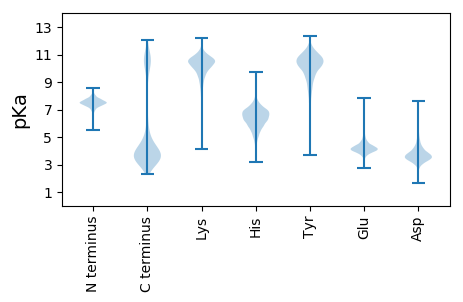
MM1 pKa = 7.26NLKK4 pKa = 10.33LLAGVSAFAFASAFAQDD21 pKa = 4.32DD22 pKa = 5.02YY23 pKa = 11.37YY24 pKa = 11.72AEE26 pKa = 5.32DD27 pKa = 4.08PAQEE31 pKa = 4.02ATPVAEE37 pKa = 4.92APAAEE42 pKa = 4.73PAPSPYY48 pKa = 8.89EE49 pKa = 4.11TEE51 pKa = 4.16PVAQAPAAVAPATVAPAQNSSSLLAQLDD79 pKa = 3.86VLRR82 pKa = 11.84GNAYY86 pKa = 10.31NFVGSEE92 pKa = 3.9LAATTVADD100 pKa = 3.82QLDD103 pKa = 4.55YY104 pKa = 10.79PYY106 pKa = 11.04MMALGQFLYY115 pKa = 10.97VEE117 pKa = 4.38PTNALGYY124 pKa = 9.67VAFNKK129 pKa = 10.53GLTFYY134 pKa = 11.11GGLDD138 pKa = 3.36NSSADD143 pKa = 3.26VGLFTAGIATAGFGLGLDD161 pKa = 3.37IGLSKK166 pKa = 11.08SFVSDD171 pKa = 3.4GDD173 pKa = 3.9DD174 pKa = 3.61DD175 pKa = 4.14ATVVNAGDD183 pKa = 3.82DD184 pKa = 3.87LGVTFAMPLGAYY196 pKa = 9.65ALSLEE201 pKa = 4.64AGWLTIDD208 pKa = 5.72DD209 pKa = 4.44EE210 pKa = 4.99VSTDD214 pKa = 3.75NVEE217 pKa = 3.64NDD219 pKa = 3.09YY220 pKa = 11.29WEE222 pKa = 4.7LSLIARR228 pKa = 11.84FTNSPSATTLGWTAGALLLRR248 pKa = 11.84HH249 pKa = 6.47AEE251 pKa = 4.21TTEE254 pKa = 3.62MTMAGQSSTSASGNSRR270 pKa = 11.84VEE272 pKa = 3.21IDD274 pKa = 3.88PYY276 pKa = 11.31FNLAAQILGNDD287 pKa = 4.0MARR290 pKa = 11.84VMIGSNNTLGIQIYY304 pKa = 10.39DD305 pKa = 3.85GGDD308 pKa = 3.05AGIKK312 pKa = 9.6SHH314 pKa = 6.45TEE316 pKa = 3.26FGLQLEE322 pKa = 4.42PNIWADD328 pKa = 3.36FALNEE333 pKa = 4.02NFLFFGGVSHH343 pKa = 6.96NVLLFGYY350 pKa = 9.84EE351 pKa = 3.98SVKK354 pKa = 10.9YY355 pKa = 10.83VDD357 pKa = 3.91GDD359 pKa = 3.92KK360 pKa = 10.77ISAIQLHH367 pKa = 5.24TEE369 pKa = 3.85RR370 pKa = 11.84TRR372 pKa = 11.84AQLGLRR378 pKa = 11.84AQYY381 pKa = 11.55KK382 pKa = 8.41MVAVEE387 pKa = 4.5ASLTNEE393 pKa = 4.16VYY395 pKa = 10.0TKK397 pKa = 10.22TVAAIFNGDD406 pKa = 4.18NIVADD411 pKa = 4.64LGIFLNFF418 pKa = 3.64
MM1 pKa = 7.26NLKK4 pKa = 10.33LLAGVSAFAFASAFAQDD21 pKa = 4.32DD22 pKa = 5.02YY23 pKa = 11.37YY24 pKa = 11.72AEE26 pKa = 5.32DD27 pKa = 4.08PAQEE31 pKa = 4.02ATPVAEE37 pKa = 4.92APAAEE42 pKa = 4.73PAPSPYY48 pKa = 8.89EE49 pKa = 4.11TEE51 pKa = 4.16PVAQAPAAVAPATVAPAQNSSSLLAQLDD79 pKa = 3.86VLRR82 pKa = 11.84GNAYY86 pKa = 10.31NFVGSEE92 pKa = 3.9LAATTVADD100 pKa = 3.82QLDD103 pKa = 4.55YY104 pKa = 10.79PYY106 pKa = 11.04MMALGQFLYY115 pKa = 10.97VEE117 pKa = 4.38PTNALGYY124 pKa = 9.67VAFNKK129 pKa = 10.53GLTFYY134 pKa = 11.11GGLDD138 pKa = 3.36NSSADD143 pKa = 3.26VGLFTAGIATAGFGLGLDD161 pKa = 3.37IGLSKK166 pKa = 11.08SFVSDD171 pKa = 3.4GDD173 pKa = 3.9DD174 pKa = 3.61DD175 pKa = 4.14ATVVNAGDD183 pKa = 3.82DD184 pKa = 3.87LGVTFAMPLGAYY196 pKa = 9.65ALSLEE201 pKa = 4.64AGWLTIDD208 pKa = 5.72DD209 pKa = 4.44EE210 pKa = 4.99VSTDD214 pKa = 3.75NVEE217 pKa = 3.64NDD219 pKa = 3.09YY220 pKa = 11.29WEE222 pKa = 4.7LSLIARR228 pKa = 11.84FTNSPSATTLGWTAGALLLRR248 pKa = 11.84HH249 pKa = 6.47AEE251 pKa = 4.21TTEE254 pKa = 3.62MTMAGQSSTSASGNSRR270 pKa = 11.84VEE272 pKa = 3.21IDD274 pKa = 3.88PYY276 pKa = 11.31FNLAAQILGNDD287 pKa = 4.0MARR290 pKa = 11.84VMIGSNNTLGIQIYY304 pKa = 10.39DD305 pKa = 3.85GGDD308 pKa = 3.05AGIKK312 pKa = 9.6SHH314 pKa = 6.45TEE316 pKa = 3.26FGLQLEE322 pKa = 4.42PNIWADD328 pKa = 3.36FALNEE333 pKa = 4.02NFLFFGGVSHH343 pKa = 6.96NVLLFGYY350 pKa = 9.84EE351 pKa = 3.98SVKK354 pKa = 10.9YY355 pKa = 10.83VDD357 pKa = 3.91GDD359 pKa = 3.92KK360 pKa = 10.77ISAIQLHH367 pKa = 5.24TEE369 pKa = 3.85RR370 pKa = 11.84TRR372 pKa = 11.84AQLGLRR378 pKa = 11.84AQYY381 pKa = 11.55KK382 pKa = 8.41MVAVEE387 pKa = 4.5ASLTNEE393 pKa = 4.16VYY395 pKa = 10.0TKK397 pKa = 10.22TVAAIFNGDD406 pKa = 4.18NIVADD411 pKa = 4.64LGIFLNFF418 pKa = 3.64
Molecular weight: 44.21 kDa
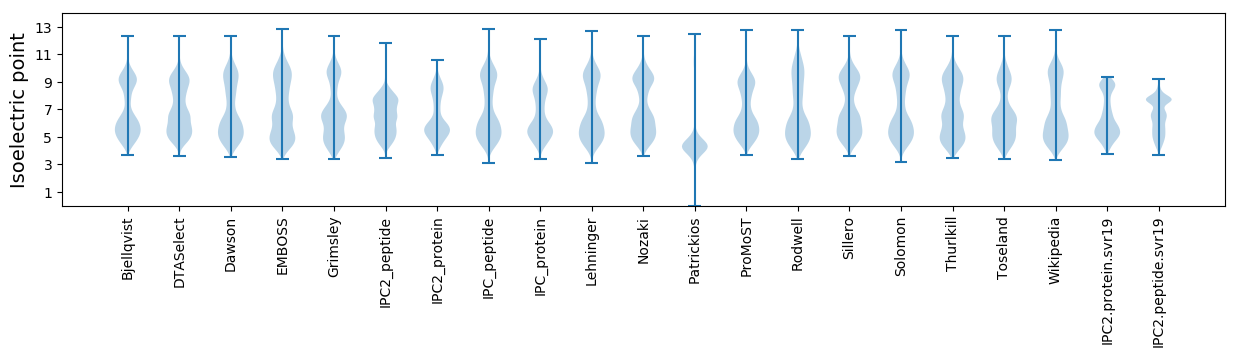
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2M9AAU5|A0A2M9AAU5_9BACT Motility quorum-sensing regulator/GCU-specific mRNA interferase toxin OS=Hallerella succinigenes OX=1896222 GN=BGX16_2781 PE=4 SV=1
MM1 pKa = 7.51GAPLRR6 pKa = 11.84RR7 pKa = 11.84GLLCFVRR14 pKa = 11.84HH15 pKa = 5.36IKK17 pKa = 9.74MYY19 pKa = 9.55FCIHH23 pKa = 5.16FRR25 pKa = 11.84IRR27 pKa = 11.84IHH29 pKa = 6.42QHH31 pKa = 4.8FQPCFSTLRR40 pKa = 11.84FLLKK44 pKa = 10.14KK45 pKa = 9.36QHH47 pKa = 6.21FPFGTGFAYY56 pKa = 10.36SCMFEE61 pKa = 4.74KK62 pKa = 10.55ISDD65 pKa = 3.83NKK67 pKa = 10.46PLTKK71 pKa = 10.24EE72 pKa = 3.74KK73 pKa = 10.67SLCSHH78 pKa = 6.52YY79 pKa = 10.76NSSSPASPPSTTSSNTSTATNNPPHH104 pKa = 7.09PNQKK108 pKa = 9.58KK109 pKa = 8.3PRR111 pKa = 11.84YY112 pKa = 7.32MRR114 pKa = 11.84NRR116 pKa = 11.84TRR118 pKa = 11.84IPRR121 pKa = 11.84QILLNPQLSYY131 pKa = 11.14LPPEE135 pKa = 4.46TNPGATRR142 pKa = 11.84IGDD145 pKa = 3.87FSLFRR150 pKa = 11.84VKK152 pKa = 10.79LCDD155 pKa = 4.23FSRR158 pKa = 11.84KK159 pKa = 8.78
MM1 pKa = 7.51GAPLRR6 pKa = 11.84RR7 pKa = 11.84GLLCFVRR14 pKa = 11.84HH15 pKa = 5.36IKK17 pKa = 9.74MYY19 pKa = 9.55FCIHH23 pKa = 5.16FRR25 pKa = 11.84IRR27 pKa = 11.84IHH29 pKa = 6.42QHH31 pKa = 4.8FQPCFSTLRR40 pKa = 11.84FLLKK44 pKa = 10.14KK45 pKa = 9.36QHH47 pKa = 6.21FPFGTGFAYY56 pKa = 10.36SCMFEE61 pKa = 4.74KK62 pKa = 10.55ISDD65 pKa = 3.83NKK67 pKa = 10.46PLTKK71 pKa = 10.24EE72 pKa = 3.74KK73 pKa = 10.67SLCSHH78 pKa = 6.52YY79 pKa = 10.76NSSSPASPPSTTSSNTSTATNNPPHH104 pKa = 7.09PNQKK108 pKa = 9.58KK109 pKa = 8.3PRR111 pKa = 11.84YY112 pKa = 7.32MRR114 pKa = 11.84NRR116 pKa = 11.84TRR118 pKa = 11.84IPRR121 pKa = 11.84QILLNPQLSYY131 pKa = 11.14LPPEE135 pKa = 4.46TNPGATRR142 pKa = 11.84IGDD145 pKa = 3.87FSLFRR150 pKa = 11.84VKK152 pKa = 10.79LCDD155 pKa = 4.23FSRR158 pKa = 11.84KK159 pKa = 8.78
Molecular weight: 18.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
925557 |
29 |
3890 |
343.6 |
38.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.316 ± 0.046 | 1.249 ± 0.018 |
5.724 ± 0.035 | 6.735 ± 0.049 |
4.853 ± 0.035 | 6.815 ± 0.05 |
1.833 ± 0.019 | 6.043 ± 0.041 |
6.771 ± 0.048 | 9.249 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.447 ± 0.021 | 4.417 ± 0.031 |
3.92 ± 0.027 | 3.075 ± 0.025 |
4.731 ± 0.037 | 6.975 ± 0.053 |
5.243 ± 0.045 | 6.748 ± 0.039 |
1.251 ± 0.017 | 3.604 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
