
Oidium neolycopersici
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Leotiomycetes; Erysiphales; Erysiphaceae; Oidium
Average proteome isoelectric point is 6.96
Get precalculated fractions of proteins
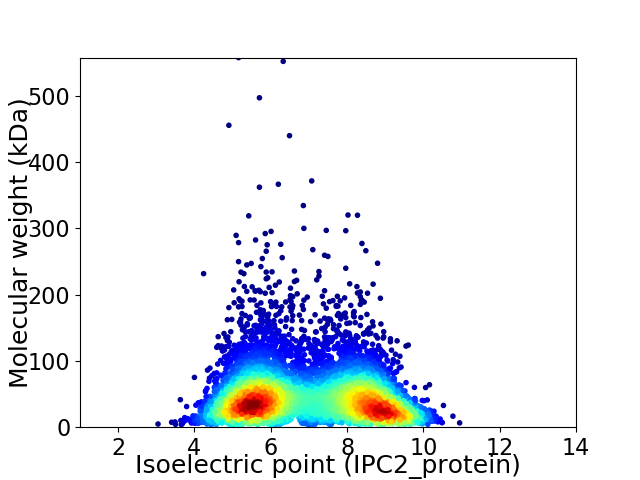
Virtual 2D-PAGE plot for 6847 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
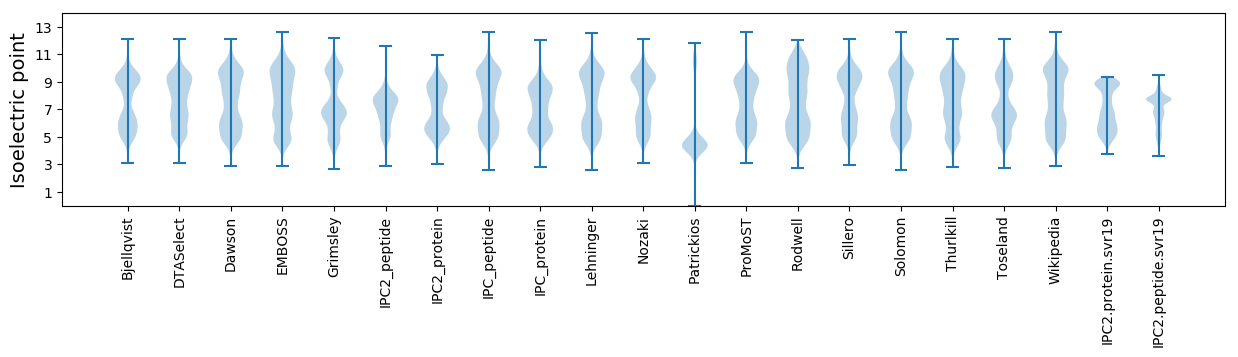
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A420HBL8|A0A420HBL8_9PEZI 3'(2') 5'-bisphosphate nucleotidase OS=Oidium neolycopersici OX=212602 GN=OnM2_094026 PE=3 SV=1
MM1 pKa = 7.56KK2 pKa = 10.54GLFAPLVAAVVLVDD16 pKa = 4.74ALTHH20 pKa = 6.0HH21 pKa = 7.32PIVYY25 pKa = 9.71YY26 pKa = 10.21PKK28 pKa = 9.1EE29 pKa = 3.85TDD31 pKa = 3.15AASLNNEE38 pKa = 4.17PGNVLKK44 pKa = 10.95SDD46 pKa = 3.43VVYY49 pKa = 11.24QNVPTNNQDD58 pKa = 3.4VEE60 pKa = 4.31PASAEE65 pKa = 3.84AVASKK70 pKa = 11.19NNISDD75 pKa = 3.51QPDD78 pKa = 3.61DD79 pKa = 4.39VDD81 pKa = 4.58EE82 pKa = 5.18LDD84 pKa = 4.42SNDD87 pKa = 3.82EE88 pKa = 4.05EE89 pKa = 6.5AEE91 pKa = 4.29TKK93 pKa = 10.43PNPTEE98 pKa = 5.19FIDD101 pKa = 5.2DD102 pKa = 3.9NQTSKK107 pKa = 9.98LTPPDD112 pKa = 3.85VKK114 pKa = 10.39PDD116 pKa = 4.34SIDD119 pKa = 4.15DD120 pKa = 4.21DD121 pKa = 4.39NLNKK125 pKa = 9.92PAGIDD130 pKa = 3.75FPDD133 pKa = 4.46GEE135 pKa = 4.25PAEE138 pKa = 4.36EE139 pKa = 4.5KK140 pKa = 10.22FDD142 pKa = 4.0SVEE145 pKa = 3.96IPNTTLYY152 pKa = 7.23TTSTVYY158 pKa = 9.74STVTSTVTDD167 pKa = 3.72CVTTVTDD174 pKa = 3.86CPEE177 pKa = 4.03IGSVVTKK184 pKa = 9.33TISMYY189 pKa = 8.68TTVCPVTTPTSVAPSSDD206 pKa = 3.04EE207 pKa = 3.94LTNEE211 pKa = 4.94DD212 pKa = 3.88SDD214 pKa = 4.92SPNSDD219 pKa = 2.15ISEE222 pKa = 4.12SDD224 pKa = 3.32EE225 pKa = 4.36PEE227 pKa = 3.77SDD229 pKa = 3.44KK230 pKa = 11.54PEE232 pKa = 4.22SDD234 pKa = 3.52EE235 pKa = 4.6PEE237 pKa = 3.9SDD239 pKa = 3.59EE240 pKa = 4.75PEE242 pKa = 4.33SEE244 pKa = 4.52SEE246 pKa = 4.45SDD248 pKa = 3.61MEE250 pKa = 5.46PDD252 pKa = 3.89TEE254 pKa = 4.92SLDD257 pKa = 3.69IPDD260 pKa = 4.58APEE263 pKa = 3.94PTQAPYY269 pKa = 8.5YY270 pKa = 8.82TKK272 pKa = 10.35TIEE275 pKa = 4.0TTVWYY280 pKa = 8.08TVTDD284 pKa = 3.98CTPKK288 pKa = 9.94EE289 pKa = 4.19TDD291 pKa = 3.66CPIGQEE297 pKa = 3.96TSSIHH302 pKa = 5.41TRR304 pKa = 11.84TEE306 pKa = 3.76IYY308 pKa = 9.96PVTDD312 pKa = 3.59IPHH315 pKa = 6.24FWSPNSTQTQPYY327 pKa = 8.16ATGSPSSYY335 pKa = 10.34PEE337 pKa = 3.74QPSNDD342 pKa = 3.14NVTIPTTPPNYY353 pKa = 9.64TINAGSAQKK362 pKa = 11.1ANALLMSFGLVAASAVAIFQGLII385 pKa = 3.31
MM1 pKa = 7.56KK2 pKa = 10.54GLFAPLVAAVVLVDD16 pKa = 4.74ALTHH20 pKa = 6.0HH21 pKa = 7.32PIVYY25 pKa = 9.71YY26 pKa = 10.21PKK28 pKa = 9.1EE29 pKa = 3.85TDD31 pKa = 3.15AASLNNEE38 pKa = 4.17PGNVLKK44 pKa = 10.95SDD46 pKa = 3.43VVYY49 pKa = 11.24QNVPTNNQDD58 pKa = 3.4VEE60 pKa = 4.31PASAEE65 pKa = 3.84AVASKK70 pKa = 11.19NNISDD75 pKa = 3.51QPDD78 pKa = 3.61DD79 pKa = 4.39VDD81 pKa = 4.58EE82 pKa = 5.18LDD84 pKa = 4.42SNDD87 pKa = 3.82EE88 pKa = 4.05EE89 pKa = 6.5AEE91 pKa = 4.29TKK93 pKa = 10.43PNPTEE98 pKa = 5.19FIDD101 pKa = 5.2DD102 pKa = 3.9NQTSKK107 pKa = 9.98LTPPDD112 pKa = 3.85VKK114 pKa = 10.39PDD116 pKa = 4.34SIDD119 pKa = 4.15DD120 pKa = 4.21DD121 pKa = 4.39NLNKK125 pKa = 9.92PAGIDD130 pKa = 3.75FPDD133 pKa = 4.46GEE135 pKa = 4.25PAEE138 pKa = 4.36EE139 pKa = 4.5KK140 pKa = 10.22FDD142 pKa = 4.0SVEE145 pKa = 3.96IPNTTLYY152 pKa = 7.23TTSTVYY158 pKa = 9.74STVTSTVTDD167 pKa = 3.72CVTTVTDD174 pKa = 3.86CPEE177 pKa = 4.03IGSVVTKK184 pKa = 9.33TISMYY189 pKa = 8.68TTVCPVTTPTSVAPSSDD206 pKa = 3.04EE207 pKa = 3.94LTNEE211 pKa = 4.94DD212 pKa = 3.88SDD214 pKa = 4.92SPNSDD219 pKa = 2.15ISEE222 pKa = 4.12SDD224 pKa = 3.32EE225 pKa = 4.36PEE227 pKa = 3.77SDD229 pKa = 3.44KK230 pKa = 11.54PEE232 pKa = 4.22SDD234 pKa = 3.52EE235 pKa = 4.6PEE237 pKa = 3.9SDD239 pKa = 3.59EE240 pKa = 4.75PEE242 pKa = 4.33SEE244 pKa = 4.52SEE246 pKa = 4.45SDD248 pKa = 3.61MEE250 pKa = 5.46PDD252 pKa = 3.89TEE254 pKa = 4.92SLDD257 pKa = 3.69IPDD260 pKa = 4.58APEE263 pKa = 3.94PTQAPYY269 pKa = 8.5YY270 pKa = 8.82TKK272 pKa = 10.35TIEE275 pKa = 4.0TTVWYY280 pKa = 8.08TVTDD284 pKa = 3.98CTPKK288 pKa = 9.94EE289 pKa = 4.19TDD291 pKa = 3.66CPIGQEE297 pKa = 3.96TSSIHH302 pKa = 5.41TRR304 pKa = 11.84TEE306 pKa = 3.76IYY308 pKa = 9.96PVTDD312 pKa = 3.59IPHH315 pKa = 6.24FWSPNSTQTQPYY327 pKa = 8.16ATGSPSSYY335 pKa = 10.34PEE337 pKa = 3.74QPSNDD342 pKa = 3.14NVTIPTTPPNYY353 pKa = 9.64TINAGSAQKK362 pKa = 11.1ANALLMSFGLVAASAVAIFQGLII385 pKa = 3.31
Molecular weight: 41.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A420HKP3|A0A420HKP3_9PEZI Putative effector protein OS=Oidium neolycopersici OX=212602 GN=OnM2_070029 PE=4 SV=1
MM1 pKa = 7.24TALLLCTSLVMLCLRR16 pKa = 11.84SRR18 pKa = 11.84SSCGGSLSRR27 pKa = 11.84IYY29 pKa = 10.74SRR31 pKa = 11.84EE32 pKa = 3.61CLARR36 pKa = 11.84SRR38 pKa = 11.84AVWHH42 pKa = 6.53ILTARR47 pKa = 11.84DD48 pKa = 3.64HH49 pKa = 6.98NITATIVLEE58 pKa = 3.78QRR60 pKa = 11.84KK61 pKa = 8.84RR62 pKa = 11.84RR63 pKa = 11.84KK64 pKa = 10.06SCDD67 pKa = 2.63AYY69 pKa = 10.65APKK72 pKa = 10.47KK73 pKa = 10.05ILQQILEE80 pKa = 4.18GPRR83 pKa = 11.84RR84 pKa = 11.84YY85 pKa = 10.23RR86 pKa = 11.84MPLGFTNKK94 pKa = 9.67RR95 pKa = 11.84QPEE98 pKa = 4.08PQISPVLHH106 pKa = 6.51EE107 pKa = 5.2FIQQ110 pKa = 3.79
MM1 pKa = 7.24TALLLCTSLVMLCLRR16 pKa = 11.84SRR18 pKa = 11.84SSCGGSLSRR27 pKa = 11.84IYY29 pKa = 10.74SRR31 pKa = 11.84EE32 pKa = 3.61CLARR36 pKa = 11.84SRR38 pKa = 11.84AVWHH42 pKa = 6.53ILTARR47 pKa = 11.84DD48 pKa = 3.64HH49 pKa = 6.98NITATIVLEE58 pKa = 3.78QRR60 pKa = 11.84KK61 pKa = 8.84RR62 pKa = 11.84RR63 pKa = 11.84KK64 pKa = 10.06SCDD67 pKa = 2.63AYY69 pKa = 10.65APKK72 pKa = 10.47KK73 pKa = 10.05ILQQILEE80 pKa = 4.18GPRR83 pKa = 11.84RR84 pKa = 11.84YY85 pKa = 10.23RR86 pKa = 11.84MPLGFTNKK94 pKa = 9.67RR95 pKa = 11.84QPEE98 pKa = 4.08PQISPVLHH106 pKa = 6.51EE107 pKa = 5.2FIQQ110 pKa = 3.79
Molecular weight: 12.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3086985 |
31 |
4886 |
450.9 |
50.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.255 ± 0.022 | 1.317 ± 0.01 |
5.43 ± 0.019 | 6.522 ± 0.031 |
3.914 ± 0.02 | 5.471 ± 0.027 |
2.244 ± 0.011 | 6.666 ± 0.022 |
6.487 ± 0.031 | 9.264 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.002 ± 0.009 | 5.224 ± 0.02 |
4.973 ± 0.022 | 4.098 ± 0.021 |
5.616 ± 0.02 | 9.369 ± 0.033 |
5.745 ± 0.015 | 5.299 ± 0.021 |
1.201 ± 0.011 | 2.901 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
