
Ophiocordyceps unilateralis
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales; Ophiocordycipitaceae; Ophiocordyceps
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
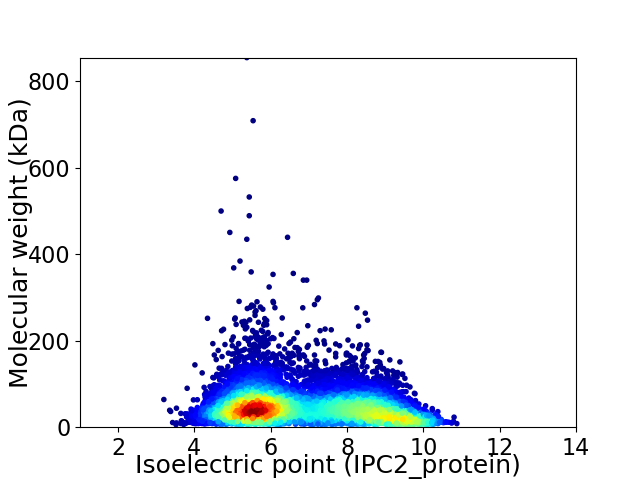
Virtual 2D-PAGE plot for 8572 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A9P7E5|A0A2A9P7E5_9HYPO Uncharacterized protein OS=Ophiocordyceps unilateralis OX=268505 GN=XA68_15485 PE=4 SV=1
MM1 pKa = 7.86KK2 pKa = 10.47YY3 pKa = 10.79NSLVLIGLAARR14 pKa = 11.84SSASPQADD22 pKa = 3.14SGAVTAKK29 pKa = 9.99IAPQGGILEE38 pKa = 4.91GCQTTYY44 pKa = 10.58PGRR47 pKa = 11.84FQVRR51 pKa = 11.84TQRR54 pKa = 11.84LSDD57 pKa = 3.51QPLVNQQPQACGADD71 pKa = 3.3KK72 pKa = 11.11SLVMTLNGGVLTDD85 pKa = 3.77GKK87 pKa = 11.28SRR89 pKa = 11.84TGYY92 pKa = 9.57VASNYY97 pKa = 8.49QFQFDD102 pKa = 4.8GPPQSGSIYY111 pKa = 8.98TAGFSLCGNGSLALGSSAVFYY132 pKa = 10.87QCLSGSFYY140 pKa = 11.4NLYY143 pKa = 10.3DD144 pKa = 4.22RR145 pKa = 11.84NWAAQCEE152 pKa = 4.98PILMMAVPCSDD163 pKa = 3.1GSGSGSAADD172 pKa = 4.1IGAGDD177 pKa = 4.11AASSSSQSPGTISQIGDD194 pKa = 3.82GQPQAPVQPQTPNQPGQAPEE214 pKa = 4.18QPVQAPNQPGQTPDD228 pKa = 3.41QPGQAPNQPGQAPEE242 pKa = 4.04QPGQGPNQPQSPVQPGAPVVSQIGDD267 pKa = 3.81GQPQAPVQPPAPVVSEE283 pKa = 4.26INDD286 pKa = 3.95GQPEE290 pKa = 4.24APAQPQTPAQPQAPTQPQAPDD311 pKa = 3.25QPQAPVVSEE320 pKa = 4.21INDD323 pKa = 3.98GQPEE327 pKa = 4.36APVGNAPPAEE337 pKa = 4.3TPATGQTPGGQSPSEE352 pKa = 4.22GGSPSQEE359 pKa = 3.6SSAPSATSAAPSSGQDD375 pKa = 2.99NTQAPEE381 pKa = 3.93ATKK384 pKa = 9.74PAYY387 pKa = 9.41EE388 pKa = 4.41VSQGMQNAASGIATALVFAMAGSFLFLL415 pKa = 4.7
MM1 pKa = 7.86KK2 pKa = 10.47YY3 pKa = 10.79NSLVLIGLAARR14 pKa = 11.84SSASPQADD22 pKa = 3.14SGAVTAKK29 pKa = 9.99IAPQGGILEE38 pKa = 4.91GCQTTYY44 pKa = 10.58PGRR47 pKa = 11.84FQVRR51 pKa = 11.84TQRR54 pKa = 11.84LSDD57 pKa = 3.51QPLVNQQPQACGADD71 pKa = 3.3KK72 pKa = 11.11SLVMTLNGGVLTDD85 pKa = 3.77GKK87 pKa = 11.28SRR89 pKa = 11.84TGYY92 pKa = 9.57VASNYY97 pKa = 8.49QFQFDD102 pKa = 4.8GPPQSGSIYY111 pKa = 8.98TAGFSLCGNGSLALGSSAVFYY132 pKa = 10.87QCLSGSFYY140 pKa = 11.4NLYY143 pKa = 10.3DD144 pKa = 4.22RR145 pKa = 11.84NWAAQCEE152 pKa = 4.98PILMMAVPCSDD163 pKa = 3.1GSGSGSAADD172 pKa = 4.1IGAGDD177 pKa = 4.11AASSSSQSPGTISQIGDD194 pKa = 3.82GQPQAPVQPQTPNQPGQAPEE214 pKa = 4.18QPVQAPNQPGQTPDD228 pKa = 3.41QPGQAPNQPGQAPEE242 pKa = 4.04QPGQGPNQPQSPVQPGAPVVSQIGDD267 pKa = 3.81GQPQAPVQPPAPVVSEE283 pKa = 4.26INDD286 pKa = 3.95GQPEE290 pKa = 4.24APAQPQTPAQPQAPTQPQAPDD311 pKa = 3.25QPQAPVVSEE320 pKa = 4.21INDD323 pKa = 3.98GQPEE327 pKa = 4.36APVGNAPPAEE337 pKa = 4.3TPATGQTPGGQSPSEE352 pKa = 4.22GGSPSQEE359 pKa = 3.6SSAPSATSAAPSSGQDD375 pKa = 2.99NTQAPEE381 pKa = 3.93ATKK384 pKa = 9.74PAYY387 pKa = 9.41EE388 pKa = 4.41VSQGMQNAASGIATALVFAMAGSFLFLL415 pKa = 4.7
Molecular weight: 41.84 kDa
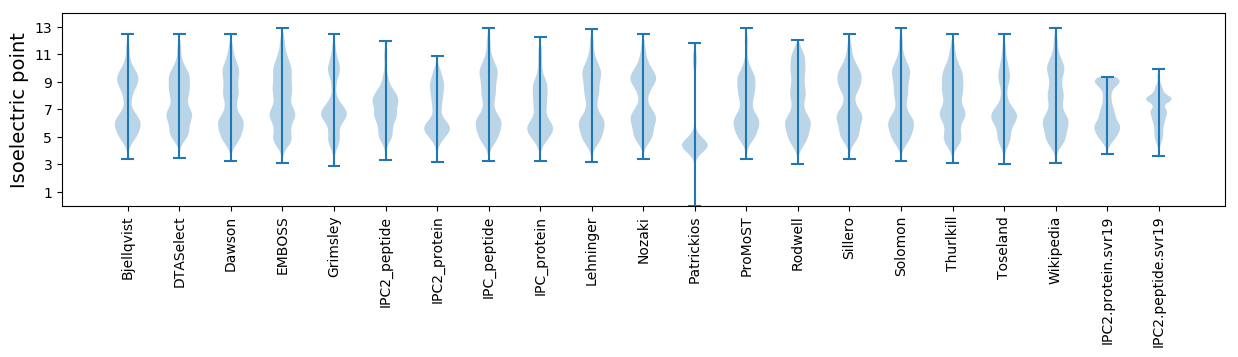
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A9PI08|A0A2A9PI08_9HYPO Uncharacterized protein OS=Ophiocordyceps unilateralis OX=268505 GN=XA68_18384 PE=4 SV=1
MM1 pKa = 7.64PCSPPLPAEE10 pKa = 4.33GKK12 pKa = 10.43LRR14 pKa = 11.84GHH16 pKa = 7.12DD17 pKa = 3.38KK18 pKa = 10.97ASNSHH23 pKa = 5.63IRR25 pKa = 11.84RR26 pKa = 11.84DD27 pKa = 3.61AIIPLRR33 pKa = 11.84QVARR37 pKa = 11.84RR38 pKa = 11.84NHH40 pKa = 5.18RR41 pKa = 11.84TGFFPVLARR50 pKa = 11.84VVYY53 pKa = 10.0SRR55 pKa = 11.84SVLALRR61 pKa = 11.84LGFCYY66 pKa = 10.3CPNVSALDD74 pKa = 3.77LLIGRR79 pKa = 11.84KK80 pKa = 8.86LWLSRR85 pKa = 11.84VGWPLCCKK93 pKa = 10.22LNNGLHH99 pKa = 5.94VGVLDD104 pKa = 4.62DD105 pKa = 4.89SSRR108 pKa = 11.84LPPLSKK114 pKa = 9.73RR115 pKa = 11.84TLIGTSASVTVEE127 pKa = 3.78KK128 pKa = 10.73KK129 pKa = 8.46HH130 pKa = 5.07THH132 pKa = 3.82THH134 pKa = 5.81TYY136 pKa = 9.78
MM1 pKa = 7.64PCSPPLPAEE10 pKa = 4.33GKK12 pKa = 10.43LRR14 pKa = 11.84GHH16 pKa = 7.12DD17 pKa = 3.38KK18 pKa = 10.97ASNSHH23 pKa = 5.63IRR25 pKa = 11.84RR26 pKa = 11.84DD27 pKa = 3.61AIIPLRR33 pKa = 11.84QVARR37 pKa = 11.84RR38 pKa = 11.84NHH40 pKa = 5.18RR41 pKa = 11.84TGFFPVLARR50 pKa = 11.84VVYY53 pKa = 10.0SRR55 pKa = 11.84SVLALRR61 pKa = 11.84LGFCYY66 pKa = 10.3CPNVSALDD74 pKa = 3.77LLIGRR79 pKa = 11.84KK80 pKa = 8.86LWLSRR85 pKa = 11.84VGWPLCCKK93 pKa = 10.22LNNGLHH99 pKa = 5.94VGVLDD104 pKa = 4.62DD105 pKa = 4.89SSRR108 pKa = 11.84LPPLSKK114 pKa = 9.73RR115 pKa = 11.84TLIGTSASVTVEE127 pKa = 3.78KK128 pKa = 10.73KK129 pKa = 8.46HH130 pKa = 5.07THH132 pKa = 3.82THH134 pKa = 5.81TYY136 pKa = 9.78
Molecular weight: 15.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4052776 |
66 |
7725 |
472.8 |
52.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.629 ± 0.027 | 1.342 ± 0.012 |
6.126 ± 0.02 | 5.789 ± 0.029 |
3.403 ± 0.017 | 7.124 ± 0.025 |
2.474 ± 0.012 | 3.907 ± 0.02 |
4.317 ± 0.023 | 9.073 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.233 ± 0.009 | 3.039 ± 0.014 |
6.409 ± 0.032 | 3.987 ± 0.021 |
7.238 ± 0.024 | 8.291 ± 0.033 |
5.46 ± 0.017 | 6.366 ± 0.019 |
1.385 ± 0.011 | 2.374 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
