
Desemzia incerta
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Carnobacteriaceae; Desemzia
Average proteome isoelectric point is 5.91
Get precalculated fractions of proteins
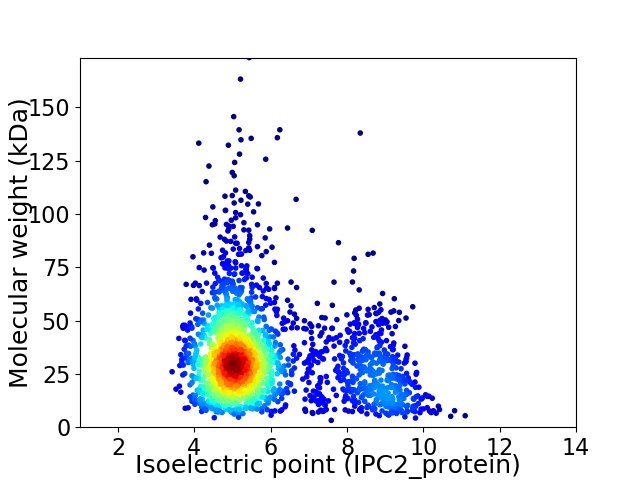
Virtual 2D-PAGE plot for 2143 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
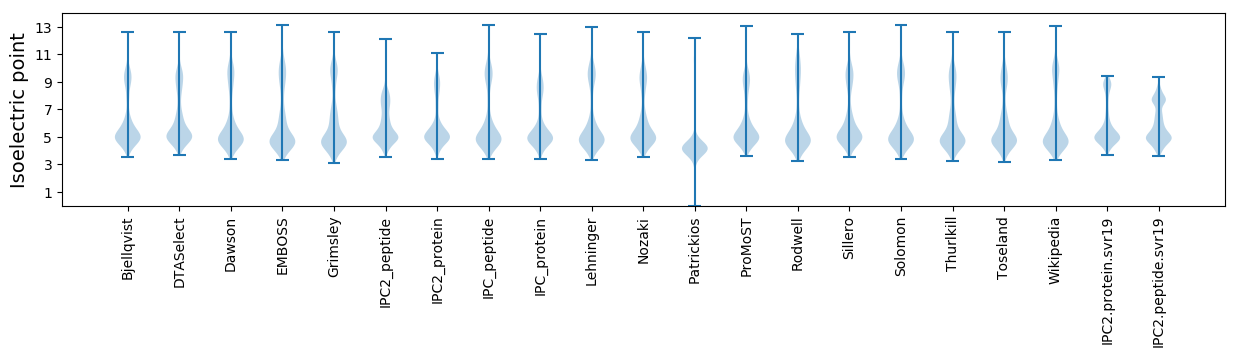
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I5WW93|A0A1I5WW93_9LACT Uncharacterized protein OS=Desemzia incerta OX=82801 GN=SAMN04488506_1142 PE=4 SV=1
MM1 pKa = 7.46NKK3 pKa = 10.22LKK5 pKa = 9.95TKK7 pKa = 10.52KK8 pKa = 10.44SFLLSLVSAGVLAGCALPGLGSGSGEE34 pKa = 3.98EE35 pKa = 4.92GITVAGVTSTEE46 pKa = 4.01GLIMSYY52 pKa = 10.01VVEE55 pKa = 4.62GMIEE59 pKa = 4.15HH60 pKa = 6.44YY61 pKa = 10.07MDD63 pKa = 5.25DD64 pKa = 5.22LDD66 pKa = 4.02VQVINNLGSSTVSHH80 pKa = 6.05QALLNGDD87 pKa = 3.9ANIAGVKK94 pKa = 8.1YY95 pKa = 9.17TGTSLTGEE103 pKa = 4.28LGEE106 pKa = 5.87DD107 pKa = 4.84PITDD111 pKa = 3.54PEE113 pKa = 4.18KK114 pKa = 10.03ALEE117 pKa = 4.23VVVEE121 pKa = 4.32GFDD124 pKa = 5.04KK125 pKa = 11.2EE126 pKa = 5.31FDD128 pKa = 4.04LKK130 pKa = 10.51WFPSYY135 pKa = 11.12GFANSYY141 pKa = 11.21AFMVTKK147 pKa = 9.23EE148 pKa = 3.91TAEE151 pKa = 4.02EE152 pKa = 4.14YY153 pKa = 10.69GLEE156 pKa = 4.46KK157 pKa = 10.41ISDD160 pKa = 4.15LEE162 pKa = 4.33PYY164 pKa = 9.75ALEE167 pKa = 4.13MNAGVDD173 pKa = 3.47NSWIEE178 pKa = 4.06RR179 pKa = 11.84EE180 pKa = 3.92GDD182 pKa = 3.55GYY184 pKa = 11.29DD185 pKa = 4.46AFLDD189 pKa = 3.92TYY191 pKa = 11.12GFDD194 pKa = 3.99FNRR197 pKa = 11.84VYY199 pKa = 10.91PMQIGLVYY207 pKa = 10.6DD208 pKa = 4.24ALQAGSMDD216 pKa = 4.14IILGYY221 pKa = 8.9STDD224 pKa = 3.62GRR226 pKa = 11.84IASYY230 pKa = 11.21DD231 pKa = 3.78LVVLEE236 pKa = 6.01DD237 pKa = 4.68DD238 pKa = 4.35LQLFPPYY245 pKa = 9.88DD246 pKa = 4.19ASPLATNEE254 pKa = 3.66ILEE257 pKa = 4.63TYY259 pKa = 10.09PEE261 pKa = 4.36LEE263 pKa = 5.11GILLRR268 pKa = 11.84MEE270 pKa = 4.15GMISEE275 pKa = 4.35EE276 pKa = 3.96QMQEE280 pKa = 3.65MNYY283 pKa = 10.31EE284 pKa = 3.82ADD286 pKa = 3.84NNLKK290 pKa = 9.89EE291 pKa = 4.43PKK293 pKa = 10.14VVADD297 pKa = 6.25DD298 pKa = 3.96FLEE301 pKa = 4.58DD302 pKa = 3.19NNYY305 pKa = 10.02FEE307 pKa = 6.25DD308 pKa = 4.76GSSNTEE314 pKa = 3.64GGEE317 pKa = 3.95
MM1 pKa = 7.46NKK3 pKa = 10.22LKK5 pKa = 9.95TKK7 pKa = 10.52KK8 pKa = 10.44SFLLSLVSAGVLAGCALPGLGSGSGEE34 pKa = 3.98EE35 pKa = 4.92GITVAGVTSTEE46 pKa = 4.01GLIMSYY52 pKa = 10.01VVEE55 pKa = 4.62GMIEE59 pKa = 4.15HH60 pKa = 6.44YY61 pKa = 10.07MDD63 pKa = 5.25DD64 pKa = 5.22LDD66 pKa = 4.02VQVINNLGSSTVSHH80 pKa = 6.05QALLNGDD87 pKa = 3.9ANIAGVKK94 pKa = 8.1YY95 pKa = 9.17TGTSLTGEE103 pKa = 4.28LGEE106 pKa = 5.87DD107 pKa = 4.84PITDD111 pKa = 3.54PEE113 pKa = 4.18KK114 pKa = 10.03ALEE117 pKa = 4.23VVVEE121 pKa = 4.32GFDD124 pKa = 5.04KK125 pKa = 11.2EE126 pKa = 5.31FDD128 pKa = 4.04LKK130 pKa = 10.51WFPSYY135 pKa = 11.12GFANSYY141 pKa = 11.21AFMVTKK147 pKa = 9.23EE148 pKa = 3.91TAEE151 pKa = 4.02EE152 pKa = 4.14YY153 pKa = 10.69GLEE156 pKa = 4.46KK157 pKa = 10.41ISDD160 pKa = 4.15LEE162 pKa = 4.33PYY164 pKa = 9.75ALEE167 pKa = 4.13MNAGVDD173 pKa = 3.47NSWIEE178 pKa = 4.06RR179 pKa = 11.84EE180 pKa = 3.92GDD182 pKa = 3.55GYY184 pKa = 11.29DD185 pKa = 4.46AFLDD189 pKa = 3.92TYY191 pKa = 11.12GFDD194 pKa = 3.99FNRR197 pKa = 11.84VYY199 pKa = 10.91PMQIGLVYY207 pKa = 10.6DD208 pKa = 4.24ALQAGSMDD216 pKa = 4.14IILGYY221 pKa = 8.9STDD224 pKa = 3.62GRR226 pKa = 11.84IASYY230 pKa = 11.21DD231 pKa = 3.78LVVLEE236 pKa = 6.01DD237 pKa = 4.68DD238 pKa = 4.35LQLFPPYY245 pKa = 9.88DD246 pKa = 4.19ASPLATNEE254 pKa = 3.66ILEE257 pKa = 4.63TYY259 pKa = 10.09PEE261 pKa = 4.36LEE263 pKa = 5.11GILLRR268 pKa = 11.84MEE270 pKa = 4.15GMISEE275 pKa = 4.35EE276 pKa = 3.96QMQEE280 pKa = 3.65MNYY283 pKa = 10.31EE284 pKa = 3.82ADD286 pKa = 3.84NNLKK290 pKa = 9.89EE291 pKa = 4.43PKK293 pKa = 10.14VVADD297 pKa = 6.25DD298 pKa = 3.96FLEE301 pKa = 4.58DD302 pKa = 3.19NNYY305 pKa = 10.02FEE307 pKa = 6.25DD308 pKa = 4.76GSSNTEE314 pKa = 3.64GGEE317 pKa = 3.95
Molecular weight: 34.7 kDa
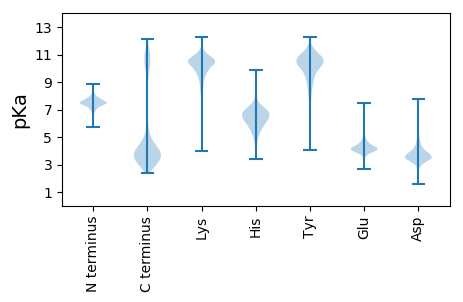
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I5XFU5|A0A1I5XFU5_9LACT Inosine-5'-monophosphate dehydrogenase OS=Desemzia incerta OX=82801 GN=guaB PE=3 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.7QPKK8 pKa = 8.83KK9 pKa = 9.05RR10 pKa = 11.84KK11 pKa = 7.43RR12 pKa = 11.84QKK14 pKa = 8.87VHH16 pKa = 5.8GFRR19 pKa = 11.84KK20 pKa = 10.04RR21 pKa = 11.84MSTKK25 pKa = 9.83NGRR28 pKa = 11.84NVLQSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.58GRR39 pKa = 11.84KK40 pKa = 8.76VLSAA44 pKa = 4.05
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.7QPKK8 pKa = 8.83KK9 pKa = 9.05RR10 pKa = 11.84KK11 pKa = 7.43RR12 pKa = 11.84QKK14 pKa = 8.87VHH16 pKa = 5.8GFRR19 pKa = 11.84KK20 pKa = 10.04RR21 pKa = 11.84MSTKK25 pKa = 9.83NGRR28 pKa = 11.84NVLQSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.58GRR39 pKa = 11.84KK40 pKa = 8.76VLSAA44 pKa = 4.05
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
661021 |
27 |
1518 |
308.5 |
34.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.104 ± 0.054 | 0.493 ± 0.012 |
5.4 ± 0.041 | 8.046 ± 0.065 |
4.39 ± 0.041 | 6.696 ± 0.056 |
1.841 ± 0.02 | 7.622 ± 0.05 |
6.593 ± 0.049 | 9.636 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.771 ± 0.021 | 4.551 ± 0.032 |
3.463 ± 0.024 | 3.912 ± 0.039 |
3.728 ± 0.036 | 6.17 ± 0.037 |
5.891 ± 0.033 | 7.156 ± 0.04 |
0.881 ± 0.018 | 3.656 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
