
Tenacibaculum adriaticum
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Tenacibaculum
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
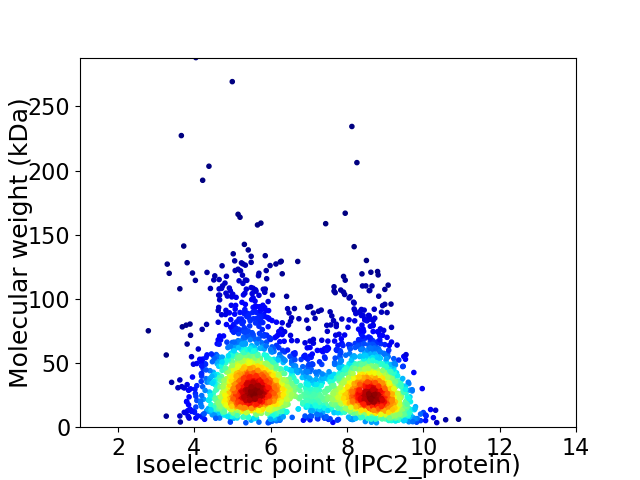
Virtual 2D-PAGE plot for 2925 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5S5DUB3|A0A5S5DUB3_9FLAO DNA repair protein RecN OS=Tenacibaculum adriaticum OX=413713 GN=C7447_102697 PE=3 SV=1
MM1 pKa = 7.27NLIKK5 pKa = 10.45KK6 pKa = 10.4SSIQLIIIILFISITACNGDD26 pKa = 3.97DD27 pKa = 5.44DD28 pKa = 6.4DD29 pKa = 7.13DD30 pKa = 4.93EE31 pKa = 5.01FGNWVEE37 pKa = 4.53SSIFDD42 pKa = 3.62GDD44 pKa = 3.39SRR46 pKa = 11.84ANAVSFTIGTNGYY59 pKa = 9.86LVTGYY64 pKa = 10.78DD65 pKa = 3.58GDD67 pKa = 5.54DD68 pKa = 3.69YY69 pKa = 11.84LSDD72 pKa = 2.86TWEE75 pKa = 4.16YY76 pKa = 11.42NSEE79 pKa = 3.74DD80 pKa = 4.42DD81 pKa = 4.01YY82 pKa = 10.15WTKK85 pKa = 10.83KK86 pKa = 10.76ADD88 pKa = 3.74FPGIARR94 pKa = 11.84SGAVGFSIDD103 pKa = 3.16GKK105 pKa = 10.63GYY107 pKa = 10.61LGTGYY112 pKa = 10.67DD113 pKa = 5.55GEE115 pKa = 5.73DD116 pKa = 3.59EE117 pKa = 6.12LNDD120 pKa = 3.45FWEE123 pKa = 4.51YY124 pKa = 11.64NPDD127 pKa = 3.47TDD129 pKa = 4.74SWTQKK134 pKa = 11.08ADD136 pKa = 3.5FGGTARR142 pKa = 11.84YY143 pKa = 8.63GAIGFSLNNDD153 pKa = 3.52GYY155 pKa = 10.59IGTGYY160 pKa = 10.59DD161 pKa = 3.23GSEE164 pKa = 3.78QKK166 pKa = 11.07DD167 pKa = 3.41FWKK170 pKa = 11.17YY171 pKa = 8.7NTTTDD176 pKa = 2.32TWEE179 pKa = 3.94QVVGFGGQKK188 pKa = 10.22RR189 pKa = 11.84KK190 pKa = 9.78DD191 pKa = 3.35ASVFTIDD198 pKa = 3.22NIAYY202 pKa = 9.46IGLGIHH208 pKa = 6.09NGSYY212 pKa = 11.0EE213 pKa = 3.98EE214 pKa = 4.84DD215 pKa = 4.02FYY217 pKa = 12.0SFDD220 pKa = 3.24GSIWTRR226 pKa = 11.84LSDD229 pKa = 5.63LDD231 pKa = 6.01DD232 pKa = 6.65DD233 pKa = 6.52DD234 pKa = 7.75DD235 pKa = 7.54DD236 pKa = 7.25DD237 pKa = 6.78DD238 pKa = 6.67DD239 pKa = 5.68YY240 pKa = 11.91EE241 pKa = 5.08ILLSSGVGFSINGKK255 pKa = 10.11GYY257 pKa = 8.19ITTGVSGSITTEE269 pKa = 3.28SWEE272 pKa = 4.22YY273 pKa = 10.49TPSTDD278 pKa = 2.79TWEE281 pKa = 4.08EE282 pKa = 3.83LPAFEE287 pKa = 5.74GSARR291 pKa = 11.84QNASAFTFATSAYY304 pKa = 9.65VLMGRR309 pKa = 11.84SGSYY313 pKa = 10.52YY314 pKa = 10.31FDD316 pKa = 4.55DD317 pKa = 3.26VWEE320 pKa = 4.43FRR322 pKa = 11.84PDD324 pKa = 3.3EE325 pKa = 4.77LEE327 pKa = 4.5NEE329 pKa = 4.27DD330 pKa = 4.74DD331 pKa = 3.7
MM1 pKa = 7.27NLIKK5 pKa = 10.45KK6 pKa = 10.4SSIQLIIIILFISITACNGDD26 pKa = 3.97DD27 pKa = 5.44DD28 pKa = 6.4DD29 pKa = 7.13DD30 pKa = 4.93EE31 pKa = 5.01FGNWVEE37 pKa = 4.53SSIFDD42 pKa = 3.62GDD44 pKa = 3.39SRR46 pKa = 11.84ANAVSFTIGTNGYY59 pKa = 9.86LVTGYY64 pKa = 10.78DD65 pKa = 3.58GDD67 pKa = 5.54DD68 pKa = 3.69YY69 pKa = 11.84LSDD72 pKa = 2.86TWEE75 pKa = 4.16YY76 pKa = 11.42NSEE79 pKa = 3.74DD80 pKa = 4.42DD81 pKa = 4.01YY82 pKa = 10.15WTKK85 pKa = 10.83KK86 pKa = 10.76ADD88 pKa = 3.74FPGIARR94 pKa = 11.84SGAVGFSIDD103 pKa = 3.16GKK105 pKa = 10.63GYY107 pKa = 10.61LGTGYY112 pKa = 10.67DD113 pKa = 5.55GEE115 pKa = 5.73DD116 pKa = 3.59EE117 pKa = 6.12LNDD120 pKa = 3.45FWEE123 pKa = 4.51YY124 pKa = 11.64NPDD127 pKa = 3.47TDD129 pKa = 4.74SWTQKK134 pKa = 11.08ADD136 pKa = 3.5FGGTARR142 pKa = 11.84YY143 pKa = 8.63GAIGFSLNNDD153 pKa = 3.52GYY155 pKa = 10.59IGTGYY160 pKa = 10.59DD161 pKa = 3.23GSEE164 pKa = 3.78QKK166 pKa = 11.07DD167 pKa = 3.41FWKK170 pKa = 11.17YY171 pKa = 8.7NTTTDD176 pKa = 2.32TWEE179 pKa = 3.94QVVGFGGQKK188 pKa = 10.22RR189 pKa = 11.84KK190 pKa = 9.78DD191 pKa = 3.35ASVFTIDD198 pKa = 3.22NIAYY202 pKa = 9.46IGLGIHH208 pKa = 6.09NGSYY212 pKa = 11.0EE213 pKa = 3.98EE214 pKa = 4.84DD215 pKa = 4.02FYY217 pKa = 12.0SFDD220 pKa = 3.24GSIWTRR226 pKa = 11.84LSDD229 pKa = 5.63LDD231 pKa = 6.01DD232 pKa = 6.65DD233 pKa = 6.52DD234 pKa = 7.75DD235 pKa = 7.54DD236 pKa = 7.25DD237 pKa = 6.78DD238 pKa = 6.67DD239 pKa = 5.68YY240 pKa = 11.91EE241 pKa = 5.08ILLSSGVGFSINGKK255 pKa = 10.11GYY257 pKa = 8.19ITTGVSGSITTEE269 pKa = 3.28SWEE272 pKa = 4.22YY273 pKa = 10.49TPSTDD278 pKa = 2.79TWEE281 pKa = 4.08EE282 pKa = 3.83LPAFEE287 pKa = 5.74GSARR291 pKa = 11.84QNASAFTFATSAYY304 pKa = 9.65VLMGRR309 pKa = 11.84SGSYY313 pKa = 10.52YY314 pKa = 10.31FDD316 pKa = 4.55DD317 pKa = 3.26VWEE320 pKa = 4.43FRR322 pKa = 11.84PDD324 pKa = 3.3EE325 pKa = 4.77LEE327 pKa = 4.5NEE329 pKa = 4.27DD330 pKa = 4.74DD331 pKa = 3.7
Molecular weight: 36.83 kDa
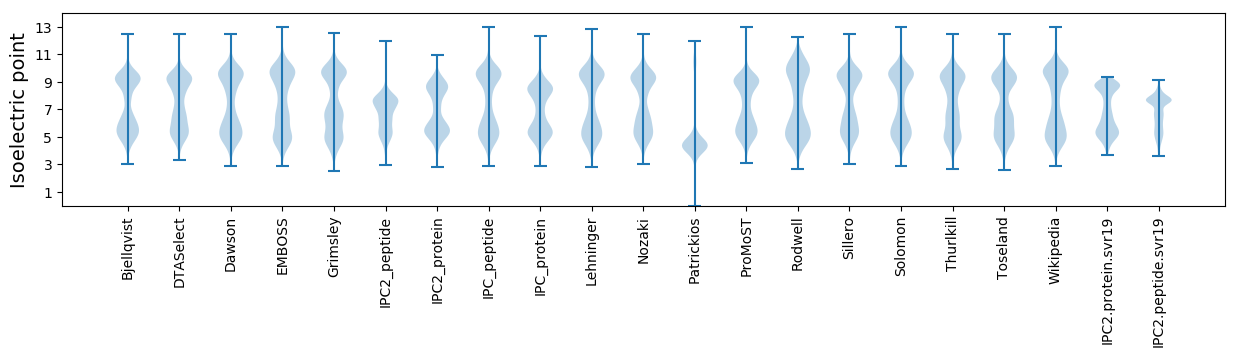
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5S5DTX7|A0A5S5DTX7_9FLAO Phytoene/squalene synthetase OS=Tenacibaculum adriaticum OX=413713 GN=C7447_102683 PE=4 SV=1
MM1 pKa = 8.03PKK3 pKa = 8.97RR4 pKa = 11.84TYY6 pKa = 10.33QPSKK10 pKa = 9.01RR11 pKa = 11.84KK12 pKa = 9.47RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.04GRR40 pKa = 11.84KK41 pKa = 8.64SLTVSSAPRR50 pKa = 11.84PKK52 pKa = 10.38KK53 pKa = 10.5
MM1 pKa = 8.03PKK3 pKa = 8.97RR4 pKa = 11.84TYY6 pKa = 10.33QPSKK10 pKa = 9.01RR11 pKa = 11.84KK12 pKa = 9.47RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.04GRR40 pKa = 11.84KK41 pKa = 8.64SLTVSSAPRR50 pKa = 11.84PKK52 pKa = 10.38KK53 pKa = 10.5
Molecular weight: 6.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
971420 |
30 |
2624 |
332.1 |
37.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.896 ± 0.047 | 0.696 ± 0.014 |
5.402 ± 0.042 | 6.744 ± 0.044 |
5.376 ± 0.039 | 6.203 ± 0.046 |
1.699 ± 0.022 | 8.385 ± 0.049 |
8.613 ± 0.056 | 9.228 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.074 ± 0.021 | 6.54 ± 0.052 |
3.17 ± 0.023 | 3.311 ± 0.024 |
3.181 ± 0.031 | 6.499 ± 0.038 |
5.821 ± 0.046 | 6.182 ± 0.039 |
0.979 ± 0.016 | 3.999 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
