
Caldivirga maquilingensis (strain ATCC 700844 / DSM 13496 / JCM 10307 / IC-167)
Taxonomy: cellular organisms; Archaea; TACK group; Crenarchaeota; Thermoprotei; Thermoproteales; Thermoproteaceae; Caldivirga; Caldivirga maquilingensis
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
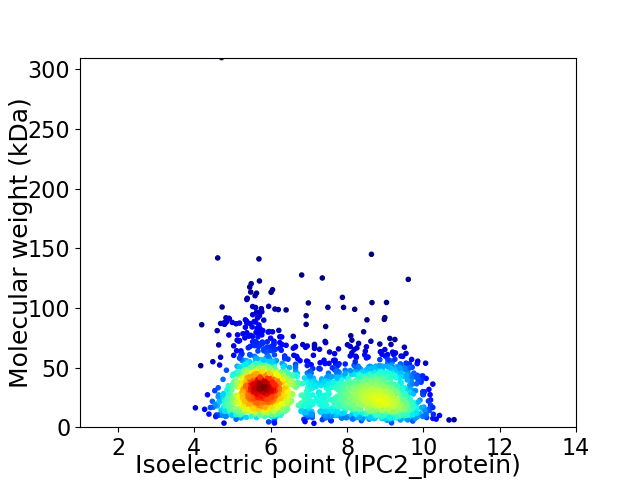
Virtual 2D-PAGE plot for 1962 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A8ME74|A8ME74_CALMQ Transcriptional regulator TrmB OS=Caldivirga maquilingensis (strain ATCC 700844 / DSM 13496 / JCM 10307 / IC-167) OX=397948 GN=Cmaq_1253 PE=4 SV=1
MM1 pKa = 7.78GIMLTRR7 pKa = 11.84LYY9 pKa = 10.84ARR11 pKa = 11.84GGITVALILAVIAIITVIMVMLIHH35 pKa = 6.91KK36 pKa = 8.88GSQPAYY42 pKa = 10.46SPVTQPITLPSGVSVLVIGPPSFVEE67 pKa = 3.9DD68 pKa = 3.34VGRR71 pKa = 11.84IIPVAEE77 pKa = 4.2SFSVDD82 pKa = 3.8SIQEE86 pKa = 4.06SPAGSVVIVDD96 pKa = 3.22WDD98 pKa = 3.85YY99 pKa = 11.84LVNQVGVKK107 pKa = 10.27GAVSDD112 pKa = 4.15LSILFKK118 pKa = 11.15QNDD121 pKa = 3.39FMIVYY126 pKa = 10.59SNDD129 pKa = 3.1SFLAAYY135 pKa = 9.12EE136 pKa = 4.22LGRR139 pKa = 11.84AWAMANGVNFTAIPAGVVGSFVAAFGDD166 pKa = 4.48SKK168 pKa = 11.24HH169 pKa = 5.6LTYY172 pKa = 10.81TSFNKK177 pKa = 8.63PTEE180 pKa = 3.98LLGAIAKK187 pKa = 9.89YY188 pKa = 10.84LEE190 pKa = 5.24LKK192 pKa = 10.56QSWQGTGQGYY202 pKa = 9.45SFSPTRR208 pKa = 11.84VLLQSNQQPTPDD220 pKa = 3.31PCYY223 pKa = 10.39QYY225 pKa = 10.31GTQEE229 pKa = 4.02GSSNGILFDD238 pKa = 4.89YY239 pKa = 10.26LPQYY243 pKa = 11.09NGVTGEE249 pKa = 4.6AYY251 pKa = 10.64SDD253 pKa = 3.89GNGTFYY259 pKa = 11.33YY260 pKa = 8.14DD261 pKa = 3.27TCIFIYY267 pKa = 10.1IYY269 pKa = 10.56GIYY272 pKa = 9.42YY273 pKa = 10.27AADD276 pKa = 3.72GTPSMDD282 pKa = 3.05VVPAVWIAYY291 pKa = 8.44VPSSTMVNNGGYY303 pKa = 9.25INYY306 pKa = 9.44YY307 pKa = 9.91VGTIDD312 pKa = 4.39HH313 pKa = 6.6EE314 pKa = 4.67KK315 pKa = 10.61GYY317 pKa = 10.29EE318 pKa = 3.9AYY320 pKa = 10.04EE321 pKa = 3.83EE322 pKa = 5.45GITNSHH328 pKa = 6.59IDD330 pKa = 3.39FAGGYY335 pKa = 9.19SPGSASGEE343 pKa = 4.17SPLTVNINYY352 pKa = 8.1EE353 pKa = 4.17NTGGATVNNTWTFTFSGYY371 pKa = 9.87PNAGQTYY378 pKa = 8.67SVAYY382 pKa = 8.94MDD384 pKa = 4.89DD385 pKa = 4.08SAWMLTQGTNNEE397 pKa = 3.83NTALLGNEE405 pKa = 3.65IGVNLVTSAQYY416 pKa = 9.26YY417 pKa = 8.78VCFDD421 pKa = 3.4ILNYY425 pKa = 10.72EE426 pKa = 4.01YY427 pKa = 10.16MYY429 pKa = 11.71VNFTWLLIYY438 pKa = 10.91NPGQKK443 pKa = 8.21PTYY446 pKa = 10.02NATTHH451 pKa = 6.51PTTSDD456 pKa = 3.28PFITGITSYY465 pKa = 10.47SQYY468 pKa = 10.88IPIPCIGG475 pKa = 3.38
MM1 pKa = 7.78GIMLTRR7 pKa = 11.84LYY9 pKa = 10.84ARR11 pKa = 11.84GGITVALILAVIAIITVIMVMLIHH35 pKa = 6.91KK36 pKa = 8.88GSQPAYY42 pKa = 10.46SPVTQPITLPSGVSVLVIGPPSFVEE67 pKa = 3.9DD68 pKa = 3.34VGRR71 pKa = 11.84IIPVAEE77 pKa = 4.2SFSVDD82 pKa = 3.8SIQEE86 pKa = 4.06SPAGSVVIVDD96 pKa = 3.22WDD98 pKa = 3.85YY99 pKa = 11.84LVNQVGVKK107 pKa = 10.27GAVSDD112 pKa = 4.15LSILFKK118 pKa = 11.15QNDD121 pKa = 3.39FMIVYY126 pKa = 10.59SNDD129 pKa = 3.1SFLAAYY135 pKa = 9.12EE136 pKa = 4.22LGRR139 pKa = 11.84AWAMANGVNFTAIPAGVVGSFVAAFGDD166 pKa = 4.48SKK168 pKa = 11.24HH169 pKa = 5.6LTYY172 pKa = 10.81TSFNKK177 pKa = 8.63PTEE180 pKa = 3.98LLGAIAKK187 pKa = 9.89YY188 pKa = 10.84LEE190 pKa = 5.24LKK192 pKa = 10.56QSWQGTGQGYY202 pKa = 9.45SFSPTRR208 pKa = 11.84VLLQSNQQPTPDD220 pKa = 3.31PCYY223 pKa = 10.39QYY225 pKa = 10.31GTQEE229 pKa = 4.02GSSNGILFDD238 pKa = 4.89YY239 pKa = 10.26LPQYY243 pKa = 11.09NGVTGEE249 pKa = 4.6AYY251 pKa = 10.64SDD253 pKa = 3.89GNGTFYY259 pKa = 11.33YY260 pKa = 8.14DD261 pKa = 3.27TCIFIYY267 pKa = 10.1IYY269 pKa = 10.56GIYY272 pKa = 9.42YY273 pKa = 10.27AADD276 pKa = 3.72GTPSMDD282 pKa = 3.05VVPAVWIAYY291 pKa = 8.44VPSSTMVNNGGYY303 pKa = 9.25INYY306 pKa = 9.44YY307 pKa = 9.91VGTIDD312 pKa = 4.39HH313 pKa = 6.6EE314 pKa = 4.67KK315 pKa = 10.61GYY317 pKa = 10.29EE318 pKa = 3.9AYY320 pKa = 10.04EE321 pKa = 3.83EE322 pKa = 5.45GITNSHH328 pKa = 6.59IDD330 pKa = 3.39FAGGYY335 pKa = 9.19SPGSASGEE343 pKa = 4.17SPLTVNINYY352 pKa = 8.1EE353 pKa = 4.17NTGGATVNNTWTFTFSGYY371 pKa = 9.87PNAGQTYY378 pKa = 8.67SVAYY382 pKa = 8.94MDD384 pKa = 4.89DD385 pKa = 4.08SAWMLTQGTNNEE397 pKa = 3.83NTALLGNEE405 pKa = 3.65IGVNLVTSAQYY416 pKa = 9.26YY417 pKa = 8.78VCFDD421 pKa = 3.4ILNYY425 pKa = 10.72EE426 pKa = 4.01YY427 pKa = 10.16MYY429 pKa = 11.71VNFTWLLIYY438 pKa = 10.91NPGQKK443 pKa = 8.21PTYY446 pKa = 10.02NATTHH451 pKa = 6.51PTTSDD456 pKa = 3.28PFITGITSYY465 pKa = 10.47SQYY468 pKa = 10.88IPIPCIGG475 pKa = 3.38
Molecular weight: 51.59 kDa
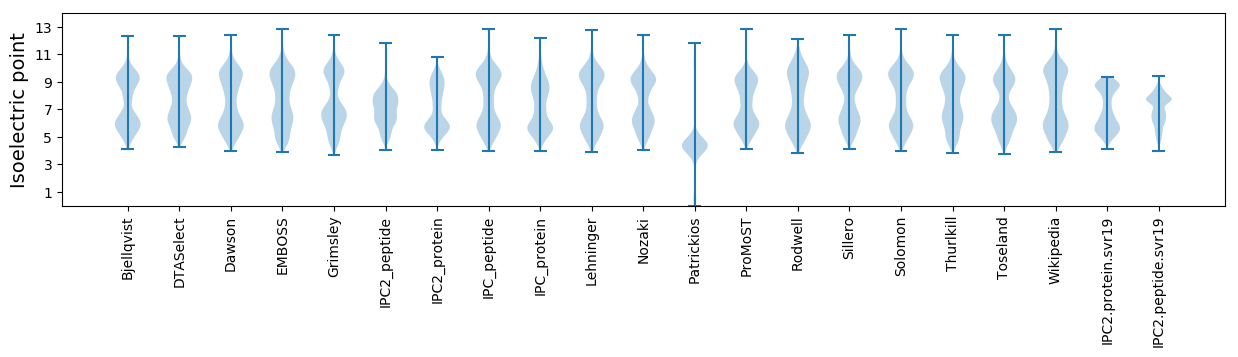
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A8MB39|A8MB39_CALMQ UspA domain protein OS=Caldivirga maquilingensis (strain ATCC 700844 / DSM 13496 / JCM 10307 / IC-167) OX=397948 GN=Cmaq_1851 PE=4 SV=1
MM1 pKa = 7.4GKK3 pKa = 9.47IKK5 pKa = 10.3PSRR8 pKa = 11.84TRR10 pKa = 11.84EE11 pKa = 3.78FGKK14 pKa = 10.82GSLRR18 pKa = 11.84CVRR21 pKa = 11.84CGTHH25 pKa = 5.25EE26 pKa = 3.99AVIRR30 pKa = 11.84RR31 pKa = 11.84YY32 pKa = 10.83GLMLCRR38 pKa = 11.84RR39 pKa = 11.84CFRR42 pKa = 11.84EE43 pKa = 3.83VAPQLGFKK51 pKa = 10.13KK52 pKa = 10.72YY53 pKa = 9.77YY54 pKa = 10.09
MM1 pKa = 7.4GKK3 pKa = 9.47IKK5 pKa = 10.3PSRR8 pKa = 11.84TRR10 pKa = 11.84EE11 pKa = 3.78FGKK14 pKa = 10.82GSLRR18 pKa = 11.84CVRR21 pKa = 11.84CGTHH25 pKa = 5.25EE26 pKa = 3.99AVIRR30 pKa = 11.84RR31 pKa = 11.84YY32 pKa = 10.83GLMLCRR38 pKa = 11.84RR39 pKa = 11.84CFRR42 pKa = 11.84EE43 pKa = 3.83VAPQLGFKK51 pKa = 10.13KK52 pKa = 10.72YY53 pKa = 9.77YY54 pKa = 10.09
Molecular weight: 6.35 kDa
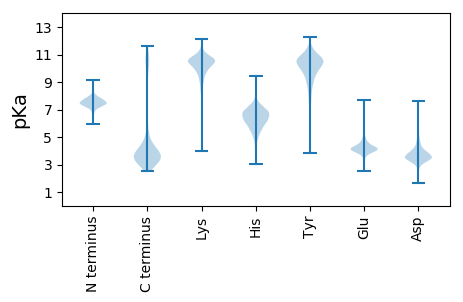
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
600938 |
30 |
2938 |
306.3 |
34.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.162 ± 0.05 | 0.652 ± 0.019 |
4.499 ± 0.044 | 5.864 ± 0.059 |
3.29 ± 0.034 | 7.56 ± 0.053 |
1.456 ± 0.022 | 8.073 ± 0.05 |
4.965 ± 0.049 | 10.676 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.65 ± 0.024 | 4.638 ± 0.046 |
4.527 ± 0.039 | 1.871 ± 0.03 |
5.664 ± 0.05 | 6.685 ± 0.049 |
5.072 ± 0.062 | 8.981 ± 0.052 |
1.32 ± 0.025 | 4.395 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
