
Amaricoccus sp. HAR-UPW-R2A-40
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Amaricoccus; unclassified Amaricoccus
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
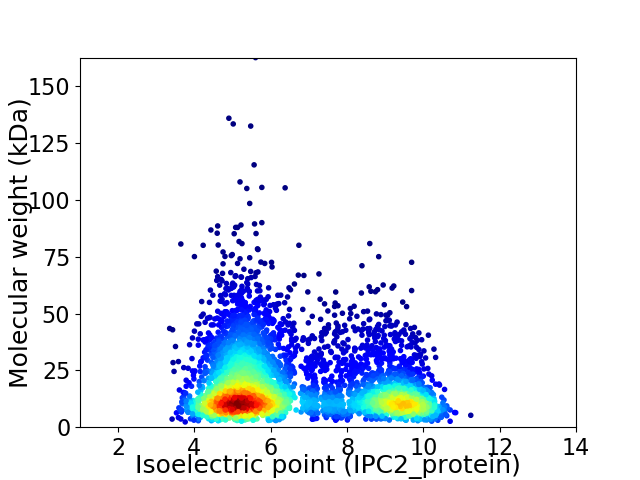
Virtual 2D-PAGE plot for 4378 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2M9PGT8|A0A2M9PGT8_9RHOB NAD(P)H dehydrogenase (quinone) OS=Amaricoccus sp. HAR-UPW-R2A-40 OX=1985299 GN=CNY89_00685 PE=3 SV=1
MM1 pKa = 7.94AYY3 pKa = 10.68LDD5 pKa = 4.08ANGVLLPTSRR15 pKa = 11.84VASSWIGGGGALYY28 pKa = 10.77GNSGDD33 pKa = 3.52NGFYY37 pKa = 10.92GSGDD41 pKa = 3.51DD42 pKa = 4.03TLTGGLGDD50 pKa = 3.99DD51 pKa = 4.65TYY53 pKa = 10.88MVWVPSTTIVEE64 pKa = 4.2AANGGVDD71 pKa = 3.58TLDD74 pKa = 3.31SRR76 pKa = 11.84VWGEE80 pKa = 4.11AILPEE85 pKa = 4.38HH86 pKa = 7.02VEE88 pKa = 3.97NLLLNGPGTTAGTGNGLRR106 pKa = 11.84NLIVAGNVGATLDD119 pKa = 4.01GLAGDD124 pKa = 4.69DD125 pKa = 4.02VLVSGAGADD134 pKa = 3.33IMRR137 pKa = 11.84VQAGNGSDD145 pKa = 3.87AIVGFVPGSDD155 pKa = 3.6VIQLVGYY162 pKa = 9.38GISTFDD168 pKa = 3.56QLAQIAAQQGSDD180 pKa = 3.87LVFTFSNDD188 pKa = 2.91EE189 pKa = 4.1KK190 pKa = 11.19LVLRR194 pKa = 11.84DD195 pKa = 3.7VVLSDD200 pKa = 4.06LDD202 pKa = 4.53GYY204 pKa = 11.41DD205 pKa = 3.77FGLDD209 pKa = 3.35QPLPPLPAGHH219 pKa = 6.28QSLFGPGQAYY229 pKa = 10.28SAFGWYY235 pKa = 9.48VLNNVWNPGPLVYY248 pKa = 10.5GVDD251 pKa = 3.89YY252 pKa = 9.83TVSSSYY258 pKa = 11.6DD259 pKa = 3.48PTDD262 pKa = 3.13LTAGVTFHH270 pKa = 6.89WAFPLTTNAFPTIIAYY286 pKa = 9.5PEE288 pKa = 4.61VIFGPAPMSGGHH300 pKa = 6.19KK301 pKa = 9.44VTDD304 pKa = 3.64TAGVFPLQVSEE315 pKa = 4.69IVDD318 pKa = 3.85LTADD322 pKa = 3.55YY323 pKa = 11.27AVAIEE328 pKa = 4.35GNTDD332 pKa = 3.36GFNVAFDD339 pKa = 3.06IWLTDD344 pKa = 3.44VPNGGPSSVTNEE356 pKa = 3.58VMVWVHH362 pKa = 6.17KK363 pKa = 11.05GGVTPYY369 pKa = 9.94GQLAGTYY376 pKa = 10.44DD377 pKa = 4.44DD378 pKa = 5.2GPVSAEE384 pKa = 4.02IYY386 pKa = 10.78VSDD389 pKa = 4.26SGDD392 pKa = 2.9WTYY395 pKa = 10.4TAVVLDD401 pKa = 3.87EE402 pKa = 4.85DD403 pKa = 4.28RR404 pKa = 11.84LVGEE408 pKa = 4.43ISVSGVLARR417 pKa = 11.84LQALGIVSSSEE428 pKa = 3.65YY429 pKa = 10.43LASLEE434 pKa = 4.18LGSEE438 pKa = 4.29IVSGAGSLTIEE449 pKa = 4.85DD450 pKa = 3.91LTLNATLEE458 pKa = 4.13DD459 pKa = 3.49RR460 pKa = 11.84TIEE463 pKa = 3.97VTGAGTTTHH472 pKa = 7.57LFPEE476 pKa = 4.99DD477 pKa = 4.07PPDD480 pKa = 3.6LSGDD484 pKa = 3.65DD485 pKa = 3.33RR486 pKa = 11.84VLYY489 pKa = 10.71DD490 pKa = 3.57PTQSLIEE497 pKa = 4.46GGEE500 pKa = 4.21GSDD503 pKa = 3.27TLVLNVGATVRR514 pKa = 11.84LDD516 pKa = 3.25RR517 pKa = 11.84FTTSQVDD524 pKa = 4.04GPAYY528 pKa = 9.22VTGFEE533 pKa = 4.43NVDD536 pKa = 3.05ASAANAGVTLYY547 pKa = 10.62GSPYY551 pKa = 10.87ANVLVGGAYY560 pKa = 9.28TDD562 pKa = 3.78TLSGGDD568 pKa = 3.51GADD571 pKa = 3.41VLRR574 pKa = 11.84GGGGGDD580 pKa = 4.01IIDD583 pKa = 4.56GGAGADD589 pKa = 3.68QIQGGDD595 pKa = 3.37GNDD598 pKa = 3.7RR599 pKa = 11.84ITYY602 pKa = 9.63DD603 pKa = 3.28AADD606 pKa = 3.63YY607 pKa = 11.18SIDD610 pKa = 3.56AGAGSDD616 pKa = 3.72TLVLTVGATVRR627 pKa = 11.84LDD629 pKa = 3.36RR630 pKa = 11.84FSTSQVDD637 pKa = 3.49GGAYY641 pKa = 7.88VTGFEE646 pKa = 4.7KK647 pKa = 10.71VDD649 pKa = 3.3AAAASAAVNLTGSAYY664 pKa = 10.56ANTLTGGSKK673 pKa = 9.99RR674 pKa = 11.84DD675 pKa = 3.68VLTGGAGADD684 pKa = 2.96QFVFKK689 pKa = 9.63TAPKK693 pKa = 10.31ASAADD698 pKa = 4.0TITDD702 pKa = 4.04FSVGEE707 pKa = 4.08DD708 pKa = 4.06RR709 pKa = 11.84IHH711 pKa = 7.34LDD713 pKa = 2.91ASFFRR718 pKa = 11.84GLPTGALASGALEE731 pKa = 4.39FGTTAAASDD740 pKa = 3.76DD741 pKa = 4.17RR742 pKa = 11.84ILYY745 pKa = 10.38DD746 pKa = 3.44SASGSLYY753 pKa = 10.37FDD755 pKa = 4.19RR756 pKa = 11.84DD757 pKa = 3.33GSADD761 pKa = 3.9DD762 pKa = 3.82YY763 pKa = 11.7SAILFATIGPGKK775 pKa = 10.13AVSAQDD781 pKa = 3.03FWVIAA786 pKa = 3.95
MM1 pKa = 7.94AYY3 pKa = 10.68LDD5 pKa = 4.08ANGVLLPTSRR15 pKa = 11.84VASSWIGGGGALYY28 pKa = 10.77GNSGDD33 pKa = 3.52NGFYY37 pKa = 10.92GSGDD41 pKa = 3.51DD42 pKa = 4.03TLTGGLGDD50 pKa = 3.99DD51 pKa = 4.65TYY53 pKa = 10.88MVWVPSTTIVEE64 pKa = 4.2AANGGVDD71 pKa = 3.58TLDD74 pKa = 3.31SRR76 pKa = 11.84VWGEE80 pKa = 4.11AILPEE85 pKa = 4.38HH86 pKa = 7.02VEE88 pKa = 3.97NLLLNGPGTTAGTGNGLRR106 pKa = 11.84NLIVAGNVGATLDD119 pKa = 4.01GLAGDD124 pKa = 4.69DD125 pKa = 4.02VLVSGAGADD134 pKa = 3.33IMRR137 pKa = 11.84VQAGNGSDD145 pKa = 3.87AIVGFVPGSDD155 pKa = 3.6VIQLVGYY162 pKa = 9.38GISTFDD168 pKa = 3.56QLAQIAAQQGSDD180 pKa = 3.87LVFTFSNDD188 pKa = 2.91EE189 pKa = 4.1KK190 pKa = 11.19LVLRR194 pKa = 11.84DD195 pKa = 3.7VVLSDD200 pKa = 4.06LDD202 pKa = 4.53GYY204 pKa = 11.41DD205 pKa = 3.77FGLDD209 pKa = 3.35QPLPPLPAGHH219 pKa = 6.28QSLFGPGQAYY229 pKa = 10.28SAFGWYY235 pKa = 9.48VLNNVWNPGPLVYY248 pKa = 10.5GVDD251 pKa = 3.89YY252 pKa = 9.83TVSSSYY258 pKa = 11.6DD259 pKa = 3.48PTDD262 pKa = 3.13LTAGVTFHH270 pKa = 6.89WAFPLTTNAFPTIIAYY286 pKa = 9.5PEE288 pKa = 4.61VIFGPAPMSGGHH300 pKa = 6.19KK301 pKa = 9.44VTDD304 pKa = 3.64TAGVFPLQVSEE315 pKa = 4.69IVDD318 pKa = 3.85LTADD322 pKa = 3.55YY323 pKa = 11.27AVAIEE328 pKa = 4.35GNTDD332 pKa = 3.36GFNVAFDD339 pKa = 3.06IWLTDD344 pKa = 3.44VPNGGPSSVTNEE356 pKa = 3.58VMVWVHH362 pKa = 6.17KK363 pKa = 11.05GGVTPYY369 pKa = 9.94GQLAGTYY376 pKa = 10.44DD377 pKa = 4.44DD378 pKa = 5.2GPVSAEE384 pKa = 4.02IYY386 pKa = 10.78VSDD389 pKa = 4.26SGDD392 pKa = 2.9WTYY395 pKa = 10.4TAVVLDD401 pKa = 3.87EE402 pKa = 4.85DD403 pKa = 4.28RR404 pKa = 11.84LVGEE408 pKa = 4.43ISVSGVLARR417 pKa = 11.84LQALGIVSSSEE428 pKa = 3.65YY429 pKa = 10.43LASLEE434 pKa = 4.18LGSEE438 pKa = 4.29IVSGAGSLTIEE449 pKa = 4.85DD450 pKa = 3.91LTLNATLEE458 pKa = 4.13DD459 pKa = 3.49RR460 pKa = 11.84TIEE463 pKa = 3.97VTGAGTTTHH472 pKa = 7.57LFPEE476 pKa = 4.99DD477 pKa = 4.07PPDD480 pKa = 3.6LSGDD484 pKa = 3.65DD485 pKa = 3.33RR486 pKa = 11.84VLYY489 pKa = 10.71DD490 pKa = 3.57PTQSLIEE497 pKa = 4.46GGEE500 pKa = 4.21GSDD503 pKa = 3.27TLVLNVGATVRR514 pKa = 11.84LDD516 pKa = 3.25RR517 pKa = 11.84FTTSQVDD524 pKa = 4.04GPAYY528 pKa = 9.22VTGFEE533 pKa = 4.43NVDD536 pKa = 3.05ASAANAGVTLYY547 pKa = 10.62GSPYY551 pKa = 10.87ANVLVGGAYY560 pKa = 9.28TDD562 pKa = 3.78TLSGGDD568 pKa = 3.51GADD571 pKa = 3.41VLRR574 pKa = 11.84GGGGGDD580 pKa = 4.01IIDD583 pKa = 4.56GGAGADD589 pKa = 3.68QIQGGDD595 pKa = 3.37GNDD598 pKa = 3.7RR599 pKa = 11.84ITYY602 pKa = 9.63DD603 pKa = 3.28AADD606 pKa = 3.63YY607 pKa = 11.18SIDD610 pKa = 3.56AGAGSDD616 pKa = 3.72TLVLTVGATVRR627 pKa = 11.84LDD629 pKa = 3.36RR630 pKa = 11.84FSTSQVDD637 pKa = 3.49GGAYY641 pKa = 7.88VTGFEE646 pKa = 4.7KK647 pKa = 10.71VDD649 pKa = 3.3AAAASAAVNLTGSAYY664 pKa = 10.56ANTLTGGSKK673 pKa = 9.99RR674 pKa = 11.84DD675 pKa = 3.68VLTGGAGADD684 pKa = 2.96QFVFKK689 pKa = 9.63TAPKK693 pKa = 10.31ASAADD698 pKa = 4.0TITDD702 pKa = 4.04FSVGEE707 pKa = 4.08DD708 pKa = 4.06RR709 pKa = 11.84IHH711 pKa = 7.34LDD713 pKa = 2.91ASFFRR718 pKa = 11.84GLPTGALASGALEE731 pKa = 4.39FGTTAAASDD740 pKa = 3.76DD741 pKa = 4.17RR742 pKa = 11.84ILYY745 pKa = 10.38DD746 pKa = 3.44SASGSLYY753 pKa = 10.37FDD755 pKa = 4.19RR756 pKa = 11.84DD757 pKa = 3.33GSADD761 pKa = 3.9DD762 pKa = 3.82YY763 pKa = 11.7SAILFATIGPGKK775 pKa = 10.13AVSAQDD781 pKa = 3.03FWVIAA786 pKa = 3.95
Molecular weight: 80.6 kDa
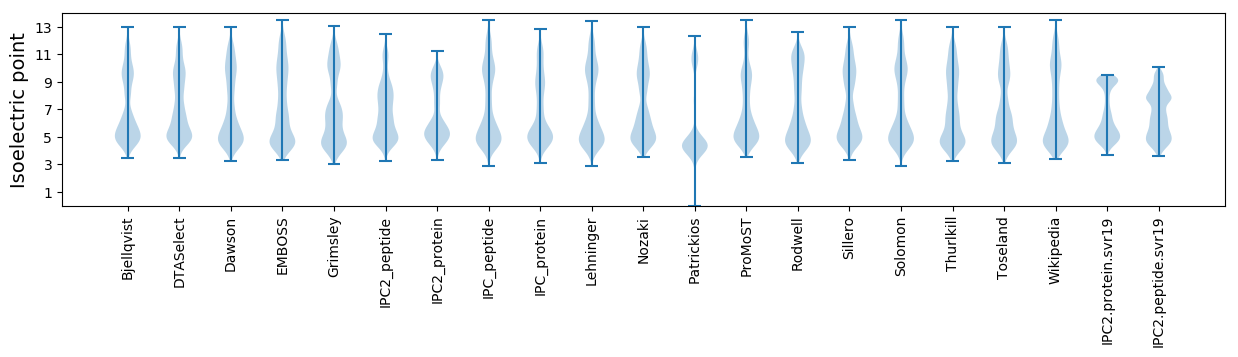
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2M9P8G0|A0A2M9P8G0_9RHOB Elongation factor Tu (Fragment) OS=Amaricoccus sp. HAR-UPW-R2A-40 OX=1985299 GN=tuf PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLVRR13 pKa = 11.84ARR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.37GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.19AGRR29 pKa = 11.84VILNRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AMGRR40 pKa = 11.84KK41 pKa = 9.03KK42 pKa = 10.63LSAA45 pKa = 3.93
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLVRR13 pKa = 11.84ARR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.37GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.19AGRR29 pKa = 11.84VILNRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AMGRR40 pKa = 11.84KK41 pKa = 9.03KK42 pKa = 10.63LSAA45 pKa = 3.93
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
800038 |
21 |
1506 |
182.7 |
19.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.635 ± 0.057 | 0.882 ± 0.015 |
5.728 ± 0.035 | 5.864 ± 0.039 |
3.841 ± 0.031 | 8.822 ± 0.046 |
2.001 ± 0.022 | 4.865 ± 0.033 |
2.824 ± 0.032 | 9.91 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.565 ± 0.02 | 2.398 ± 0.025 |
5.445 ± 0.033 | 2.942 ± 0.023 |
7.759 ± 0.06 | 5.08 ± 0.031 |
5.255 ± 0.033 | 7.392 ± 0.038 |
1.532 ± 0.021 | 2.259 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
