
SAR202 cluster bacterium AD-812-D07_MRT_10900m
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Chloroflexi incertae sedis; SAR202 cluster
Average proteome isoelectric point is 5.91
Get precalculated fractions of proteins
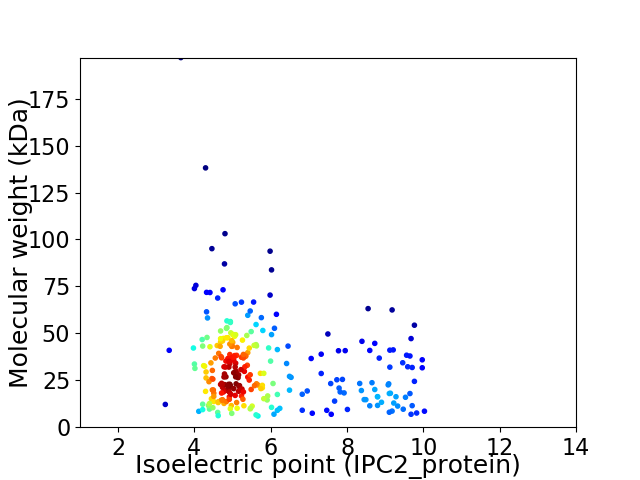
Virtual 2D-PAGE plot for 274 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5N8YXV4|A0A5N8YXV4_9CHLR Uncharacterized protein OS=SAR202 cluster bacterium AD-812-D07_MRT_10900m OX=2587835 GN=FIL92_00715 PE=4 SV=1
MM1 pKa = 7.49ACAAGITNEE10 pKa = 4.3LLVPLTGSSEE20 pKa = 4.17MFSGAKK26 pKa = 9.99NKK28 pKa = 9.28WLVLVVVAALGAFVIACGTDD48 pKa = 3.39EE49 pKa = 4.81TPVTQNDD56 pKa = 4.41GVGSQPGQNGSGGGSIIADD75 pKa = 3.59SQALEE80 pKa = 4.08TALEE84 pKa = 4.12LEE86 pKa = 4.63GADD89 pKa = 3.77VEE91 pKa = 5.14FGVQSEE97 pKa = 4.29FAGFFGLRR105 pKa = 11.84PTEE108 pKa = 3.8LKK110 pKa = 11.27VNGEE114 pKa = 3.98SLLIYY119 pKa = 9.76EE120 pKa = 4.9FAPGTSAEE128 pKa = 4.19EE129 pKa = 4.03ASEE132 pKa = 4.33GVSPDD137 pKa = 3.65GITMVNPDD145 pKa = 3.43GSATSVFWAAPPHH158 pKa = 6.65FYY160 pKa = 11.14LFGNSILLYY169 pKa = 9.89NGNDD173 pKa = 3.49AEE175 pKa = 4.49IGALLGSVSVQFAGRR190 pKa = 11.84DD191 pKa = 3.7FEE193 pKa = 4.54EE194 pKa = 4.47VSNGSGSGSGDD205 pKa = 3.5LGDD208 pKa = 5.35PPPSDD213 pKa = 3.25PGFEE217 pKa = 4.25VVEE220 pKa = 3.92ALAPIEE226 pKa = 4.17SVEE229 pKa = 3.9ILTLEE234 pKa = 4.33SYY236 pKa = 10.54PEE238 pKa = 3.78QFIVQVTSSLPNGCASYY255 pKa = 10.88SHH257 pKa = 6.92NEE259 pKa = 3.84VTQDD263 pKa = 2.98GTDD266 pKa = 3.14IKK268 pKa = 10.65ISVYY272 pKa = 9.58NTVPAPGEE280 pKa = 4.26LIACTEE286 pKa = 4.36IYY288 pKa = 10.14RR289 pKa = 11.84LHH291 pKa = 6.83DD292 pKa = 3.55QNIGLGSDD300 pKa = 4.03FEE302 pKa = 5.33RR303 pKa = 11.84GTTYY307 pKa = 10.24TVLVNDD313 pKa = 4.42HH314 pKa = 6.99PGEE317 pKa = 4.25TFTTGSAPLPSGATPPAPEE336 pKa = 4.05VPVDD340 pKa = 3.89HH341 pKa = 7.03EE342 pKa = 4.96LVTAPIEE349 pKa = 4.09SLEE352 pKa = 4.35IIQGEE357 pKa = 4.3DD358 pKa = 2.92SRR360 pKa = 11.84GRR362 pKa = 11.84ATYY365 pKa = 9.36SARR368 pKa = 11.84VAWGLSDD375 pKa = 4.3GCKK378 pKa = 9.6QSYY381 pKa = 10.13NRR383 pKa = 11.84TISRR387 pKa = 11.84IDD389 pKa = 3.22EE390 pKa = 4.11TTFEE394 pKa = 4.07IKK396 pKa = 10.53AIVTSPTGDD405 pKa = 3.4VMCTLDD411 pKa = 3.93YY412 pKa = 10.42RR413 pKa = 11.84TDD415 pKa = 3.2SDD417 pKa = 4.48DD418 pKa = 4.16FYY420 pKa = 11.49LGAVGEE426 pKa = 4.49ALTACTVYY434 pKa = 10.71HH435 pKa = 6.39IVAGKK440 pKa = 10.23LRR442 pKa = 11.84VEE444 pKa = 4.39YY445 pKa = 10.4QAIAPNVRR453 pKa = 11.84CVDD456 pKa = 3.73PEE458 pKa = 4.17LTPSAGSGGGSIIADD473 pKa = 3.64SQALEE478 pKa = 4.36LSLEE482 pKa = 4.34SKK484 pKa = 10.69GADD487 pKa = 3.33VEE489 pKa = 4.51FGGASAFSKK498 pKa = 10.76LFGVAPSEE506 pKa = 4.3LKK508 pKa = 11.27VNGQAVQIYY517 pKa = 8.76QFAPGTSAEE526 pKa = 4.22EE527 pKa = 4.14ASEE530 pKa = 4.3SVSPGGTTIVNPDD543 pKa = 3.06GSVISVMWIAPPHH556 pKa = 6.41FYY558 pKa = 11.06LFGNAIILYY567 pKa = 9.33VGNDD571 pKa = 3.65PEE573 pKa = 5.41IGALLDD579 pKa = 3.71SVAGKK584 pKa = 9.58FAGSNFEE591 pKa = 4.4DD592 pKa = 3.85AGSGSGEE599 pKa = 3.62IDD601 pKa = 3.01NEE603 pKa = 3.92YY604 pKa = 10.37RR605 pKa = 11.84IQTAQIVRR613 pKa = 11.84VDD615 pKa = 3.28IASTRR620 pKa = 11.84SIPAQHH626 pKa = 6.72MISMTIALGGSCEE639 pKa = 4.06EE640 pKa = 4.41FAGLDD645 pKa = 3.17WRR647 pKa = 11.84VEE649 pKa = 3.91GRR651 pKa = 11.84EE652 pKa = 3.93VIIDD656 pKa = 3.62VTTKK660 pKa = 10.45VPTAPVPCTLAIIYY674 pKa = 8.53EE675 pKa = 4.22DD676 pKa = 3.37QSVNIGDD683 pKa = 3.77EE684 pKa = 4.39YY685 pKa = 10.06EE686 pKa = 4.17TGVEE690 pKa = 3.64YY691 pKa = 10.86DD692 pKa = 3.63VIVNGEE698 pKa = 4.01RR699 pKa = 11.84QGTFIGGG706 pKa = 3.31
MM1 pKa = 7.49ACAAGITNEE10 pKa = 4.3LLVPLTGSSEE20 pKa = 4.17MFSGAKK26 pKa = 9.99NKK28 pKa = 9.28WLVLVVVAALGAFVIACGTDD48 pKa = 3.39EE49 pKa = 4.81TPVTQNDD56 pKa = 4.41GVGSQPGQNGSGGGSIIADD75 pKa = 3.59SQALEE80 pKa = 4.08TALEE84 pKa = 4.12LEE86 pKa = 4.63GADD89 pKa = 3.77VEE91 pKa = 5.14FGVQSEE97 pKa = 4.29FAGFFGLRR105 pKa = 11.84PTEE108 pKa = 3.8LKK110 pKa = 11.27VNGEE114 pKa = 3.98SLLIYY119 pKa = 9.76EE120 pKa = 4.9FAPGTSAEE128 pKa = 4.19EE129 pKa = 4.03ASEE132 pKa = 4.33GVSPDD137 pKa = 3.65GITMVNPDD145 pKa = 3.43GSATSVFWAAPPHH158 pKa = 6.65FYY160 pKa = 11.14LFGNSILLYY169 pKa = 9.89NGNDD173 pKa = 3.49AEE175 pKa = 4.49IGALLGSVSVQFAGRR190 pKa = 11.84DD191 pKa = 3.7FEE193 pKa = 4.54EE194 pKa = 4.47VSNGSGSGSGDD205 pKa = 3.5LGDD208 pKa = 5.35PPPSDD213 pKa = 3.25PGFEE217 pKa = 4.25VVEE220 pKa = 3.92ALAPIEE226 pKa = 4.17SVEE229 pKa = 3.9ILTLEE234 pKa = 4.33SYY236 pKa = 10.54PEE238 pKa = 3.78QFIVQVTSSLPNGCASYY255 pKa = 10.88SHH257 pKa = 6.92NEE259 pKa = 3.84VTQDD263 pKa = 2.98GTDD266 pKa = 3.14IKK268 pKa = 10.65ISVYY272 pKa = 9.58NTVPAPGEE280 pKa = 4.26LIACTEE286 pKa = 4.36IYY288 pKa = 10.14RR289 pKa = 11.84LHH291 pKa = 6.83DD292 pKa = 3.55QNIGLGSDD300 pKa = 4.03FEE302 pKa = 5.33RR303 pKa = 11.84GTTYY307 pKa = 10.24TVLVNDD313 pKa = 4.42HH314 pKa = 6.99PGEE317 pKa = 4.25TFTTGSAPLPSGATPPAPEE336 pKa = 4.05VPVDD340 pKa = 3.89HH341 pKa = 7.03EE342 pKa = 4.96LVTAPIEE349 pKa = 4.09SLEE352 pKa = 4.35IIQGEE357 pKa = 4.3DD358 pKa = 2.92SRR360 pKa = 11.84GRR362 pKa = 11.84ATYY365 pKa = 9.36SARR368 pKa = 11.84VAWGLSDD375 pKa = 4.3GCKK378 pKa = 9.6QSYY381 pKa = 10.13NRR383 pKa = 11.84TISRR387 pKa = 11.84IDD389 pKa = 3.22EE390 pKa = 4.11TTFEE394 pKa = 4.07IKK396 pKa = 10.53AIVTSPTGDD405 pKa = 3.4VMCTLDD411 pKa = 3.93YY412 pKa = 10.42RR413 pKa = 11.84TDD415 pKa = 3.2SDD417 pKa = 4.48DD418 pKa = 4.16FYY420 pKa = 11.49LGAVGEE426 pKa = 4.49ALTACTVYY434 pKa = 10.71HH435 pKa = 6.39IVAGKK440 pKa = 10.23LRR442 pKa = 11.84VEE444 pKa = 4.39YY445 pKa = 10.4QAIAPNVRR453 pKa = 11.84CVDD456 pKa = 3.73PEE458 pKa = 4.17LTPSAGSGGGSIIADD473 pKa = 3.64SQALEE478 pKa = 4.36LSLEE482 pKa = 4.34SKK484 pKa = 10.69GADD487 pKa = 3.33VEE489 pKa = 4.51FGGASAFSKK498 pKa = 10.76LFGVAPSEE506 pKa = 4.3LKK508 pKa = 11.27VNGQAVQIYY517 pKa = 8.76QFAPGTSAEE526 pKa = 4.22EE527 pKa = 4.14ASEE530 pKa = 4.3SVSPGGTTIVNPDD543 pKa = 3.06GSVISVMWIAPPHH556 pKa = 6.41FYY558 pKa = 11.06LFGNAIILYY567 pKa = 9.33VGNDD571 pKa = 3.65PEE573 pKa = 5.41IGALLDD579 pKa = 3.71SVAGKK584 pKa = 9.58FAGSNFEE591 pKa = 4.4DD592 pKa = 3.85AGSGSGEE599 pKa = 3.62IDD601 pKa = 3.01NEE603 pKa = 3.92YY604 pKa = 10.37RR605 pKa = 11.84IQTAQIVRR613 pKa = 11.84VDD615 pKa = 3.28IASTRR620 pKa = 11.84SIPAQHH626 pKa = 6.72MISMTIALGGSCEE639 pKa = 4.06EE640 pKa = 4.41FAGLDD645 pKa = 3.17WRR647 pKa = 11.84VEE649 pKa = 3.91GRR651 pKa = 11.84EE652 pKa = 3.93VIIDD656 pKa = 3.62VTTKK660 pKa = 10.45VPTAPVPCTLAIIYY674 pKa = 8.53EE675 pKa = 4.22DD676 pKa = 3.37QSVNIGDD683 pKa = 3.77EE684 pKa = 4.39YY685 pKa = 10.06EE686 pKa = 4.17TGVEE690 pKa = 3.64YY691 pKa = 10.86DD692 pKa = 3.63VIVNGEE698 pKa = 4.01RR699 pKa = 11.84QGTFIGGG706 pKa = 3.31
Molecular weight: 73.84 kDa
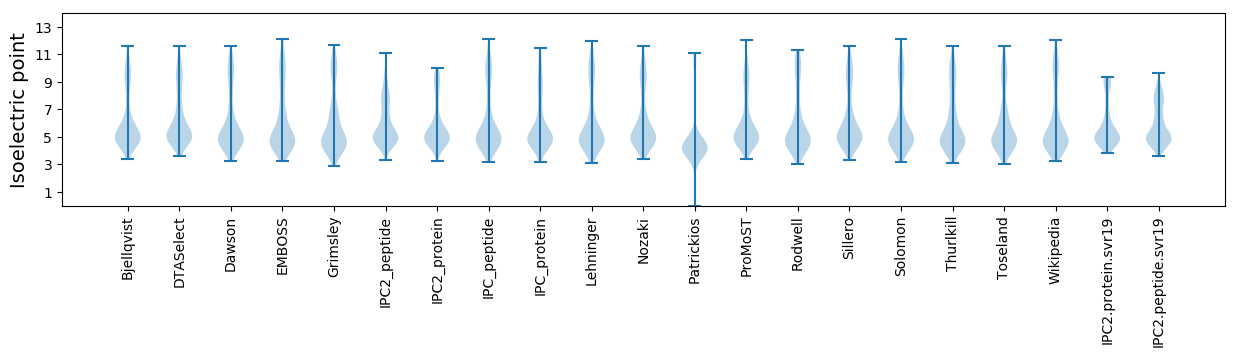
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5N8YYA7|A0A5N8YYA7_9CHLR DNA repair protein RadA OS=SAR202 cluster bacterium AD-812-D07_MRT_10900m OX=2587835 GN=radA PE=3 SV=1
MM1 pKa = 7.09EE2 pKa = 4.89WFRR5 pKa = 11.84RR6 pKa = 11.84WIVEE10 pKa = 4.41GYY12 pKa = 8.62SVRR15 pKa = 11.84QLALQSGHH23 pKa = 5.78SRR25 pKa = 11.84RR26 pKa = 11.84KK27 pKa = 9.29LRR29 pKa = 11.84RR30 pKa = 11.84LIDD33 pKa = 3.6SFLAQAPPTSALEE46 pKa = 4.13PQTARR51 pKa = 11.84YY52 pKa = 9.46LLFDD56 pKa = 3.35GTFLHH61 pKa = 6.86RR62 pKa = 11.84PHH64 pKa = 7.77SIVVLMNGQTHH75 pKa = 6.39RR76 pKa = 11.84LVRR79 pKa = 11.84GQFDD83 pKa = 3.36VRR85 pKa = 11.84EE86 pKa = 4.04NSGPQLRR93 pKa = 11.84AFFEE97 pKa = 4.23PMMDD101 pKa = 3.09EE102 pKa = 4.22GLRR105 pKa = 11.84PLSFTVDD112 pKa = 3.08GNRR115 pKa = 11.84QVIRR119 pKa = 11.84VLRR122 pKa = 11.84TLWPDD127 pKa = 3.27AVIQRR132 pKa = 11.84CLVHH136 pKa = 6.42IQRR139 pKa = 11.84QGLSWCRR146 pKa = 11.84ISPKK150 pKa = 9.85TPYY153 pKa = 10.46ARR155 pKa = 11.84QLRR158 pKa = 11.84DD159 pKa = 2.84IFLQVTKK166 pKa = 10.31IATPADD172 pKa = 3.61RR173 pKa = 11.84EE174 pKa = 4.27AFLNLVAIWEE184 pKa = 4.21EE185 pKa = 4.49RR186 pKa = 11.84YY187 pKa = 8.82GTEE190 pKa = 3.06IDD192 pKa = 3.21ARR194 pKa = 11.84KK195 pKa = 9.78EE196 pKa = 3.52IGRR199 pKa = 11.84VFSDD203 pKa = 3.27IKK205 pKa = 10.29RR206 pKa = 11.84ARR208 pKa = 11.84SMLMHH213 pKa = 7.46ALPDD217 pKa = 3.59MFHH220 pKa = 7.14YY221 pKa = 10.85LDD223 pKa = 4.92DD224 pKa = 3.81RR225 pKa = 11.84HH226 pKa = 7.25IPTTTNGLEE235 pKa = 4.65GYY237 pKa = 10.14FSRR240 pKa = 11.84LKK242 pKa = 9.89SHH244 pKa = 5.96YY245 pKa = 8.3RR246 pKa = 11.84QHH248 pKa = 7.21RR249 pKa = 11.84GLSPRR254 pKa = 11.84KK255 pKa = 9.15RR256 pKa = 11.84PNYY259 pKa = 7.99FAWYY263 pKa = 8.81FHH265 pKa = 6.28FTPKK269 pKa = 10.65
MM1 pKa = 7.09EE2 pKa = 4.89WFRR5 pKa = 11.84RR6 pKa = 11.84WIVEE10 pKa = 4.41GYY12 pKa = 8.62SVRR15 pKa = 11.84QLALQSGHH23 pKa = 5.78SRR25 pKa = 11.84RR26 pKa = 11.84KK27 pKa = 9.29LRR29 pKa = 11.84RR30 pKa = 11.84LIDD33 pKa = 3.6SFLAQAPPTSALEE46 pKa = 4.13PQTARR51 pKa = 11.84YY52 pKa = 9.46LLFDD56 pKa = 3.35GTFLHH61 pKa = 6.86RR62 pKa = 11.84PHH64 pKa = 7.77SIVVLMNGQTHH75 pKa = 6.39RR76 pKa = 11.84LVRR79 pKa = 11.84GQFDD83 pKa = 3.36VRR85 pKa = 11.84EE86 pKa = 4.04NSGPQLRR93 pKa = 11.84AFFEE97 pKa = 4.23PMMDD101 pKa = 3.09EE102 pKa = 4.22GLRR105 pKa = 11.84PLSFTVDD112 pKa = 3.08GNRR115 pKa = 11.84QVIRR119 pKa = 11.84VLRR122 pKa = 11.84TLWPDD127 pKa = 3.27AVIQRR132 pKa = 11.84CLVHH136 pKa = 6.42IQRR139 pKa = 11.84QGLSWCRR146 pKa = 11.84ISPKK150 pKa = 9.85TPYY153 pKa = 10.46ARR155 pKa = 11.84QLRR158 pKa = 11.84DD159 pKa = 2.84IFLQVTKK166 pKa = 10.31IATPADD172 pKa = 3.61RR173 pKa = 11.84EE174 pKa = 4.27AFLNLVAIWEE184 pKa = 4.21EE185 pKa = 4.49RR186 pKa = 11.84YY187 pKa = 8.82GTEE190 pKa = 3.06IDD192 pKa = 3.21ARR194 pKa = 11.84KK195 pKa = 9.78EE196 pKa = 3.52IGRR199 pKa = 11.84VFSDD203 pKa = 3.27IKK205 pKa = 10.29RR206 pKa = 11.84ARR208 pKa = 11.84SMLMHH213 pKa = 7.46ALPDD217 pKa = 3.59MFHH220 pKa = 7.14YY221 pKa = 10.85LDD223 pKa = 4.92DD224 pKa = 3.81RR225 pKa = 11.84HH226 pKa = 7.25IPTTTNGLEE235 pKa = 4.65GYY237 pKa = 10.14FSRR240 pKa = 11.84LKK242 pKa = 9.89SHH244 pKa = 5.96YY245 pKa = 8.3RR246 pKa = 11.84QHH248 pKa = 7.21RR249 pKa = 11.84GLSPRR254 pKa = 11.84KK255 pKa = 9.15RR256 pKa = 11.84PNYY259 pKa = 7.99FAWYY263 pKa = 8.81FHH265 pKa = 6.28FTPKK269 pKa = 10.65
Molecular weight: 31.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
81315 |
52 |
1923 |
296.8 |
32.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.463 ± 0.162 | 0.767 ± 0.042 |
6.328 ± 0.125 | 6.384 ± 0.129 |
3.633 ± 0.084 | 8.654 ± 0.164 |
2.019 ± 0.073 | 5.695 ± 0.086 |
3.252 ± 0.126 | 9.066 ± 0.134 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.236 ± 0.071 | 3.264 ± 0.077 |
5.115 ± 0.11 | 2.972 ± 0.074 |
6.44 ± 0.184 | 6.921 ± 0.141 |
5.987 ± 0.161 | 7.926 ± 0.127 |
1.439 ± 0.061 | 2.439 ± 0.077 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
