
Rhizopus delemar (strain RA 99-880 / ATCC MYA-4621 / FGSC 9543 / NRRL 43880) (Mucormycosis agent) (Rhizopus arrhizus var. delemar)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Mucoromycota; Mucoromycotina; Mucoromycetes; Mucorales; Mucorineae; Rhizopodaceae; Rhizopus; Rhizopus delemar
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
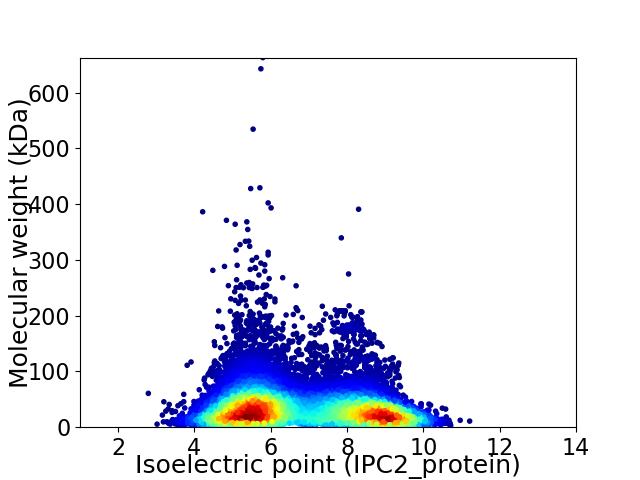
Virtual 2D-PAGE plot for 16971 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I1BU05|I1BU05_RHIO9 Uncharacterized protein OS=Rhizopus delemar (strain RA 99-880 / ATCC MYA-4621 / FGSC 9543 / NRRL 43880) OX=246409 GN=RO3G_04390 PE=4 SV=1
MM1 pKa = 7.42YY2 pKa = 9.71YY3 pKa = 10.73SSITIISSILLATCSFVNAASTNSSYY29 pKa = 11.41PLTTINSQKK38 pKa = 10.41DD39 pKa = 3.29FCIFLPPQPGLEE51 pKa = 4.08VAVNEE56 pKa = 4.19NNGIPFCFNQDD67 pKa = 3.19AVPNATVFPTDD78 pKa = 4.72FITTAHH84 pKa = 5.95YY85 pKa = 10.57LKK87 pKa = 9.98TSTYY91 pKa = 9.02VQITGFFDD99 pKa = 3.13RR100 pKa = 11.84TKK102 pKa = 11.2YY103 pKa = 10.51DD104 pKa = 3.34LQEE107 pKa = 4.09TDD109 pKa = 4.72GGGQYY114 pKa = 10.89DD115 pKa = 3.6NHH117 pKa = 6.84AKK119 pKa = 10.07GKK121 pKa = 9.46PVGAQCKK128 pKa = 9.12GYY130 pKa = 10.66NYY132 pKa = 8.96FVNMIEE138 pKa = 4.38PDD140 pKa = 3.41LQRR143 pKa = 11.84FCIRR147 pKa = 11.84CCQDD151 pKa = 3.97KK152 pKa = 10.97DD153 pKa = 3.57DD154 pKa = 5.29CNTGRR159 pKa = 11.84SGYY162 pKa = 9.87GCLRR166 pKa = 11.84IITGDD171 pKa = 3.59YY172 pKa = 10.55TDD174 pKa = 4.86DD175 pKa = 3.59NNMIGNSTTSSGHH188 pKa = 5.7SNNHH192 pKa = 5.43INSVLAEE199 pKa = 4.21LDD201 pKa = 3.83EE202 pKa = 5.16LPAATDD208 pKa = 3.44STNATDD214 pKa = 3.64STNATDD220 pKa = 3.86STDD223 pKa = 3.39TATSADD229 pKa = 3.88TADD232 pKa = 3.79SSSTEE237 pKa = 4.19SNSALDD243 pKa = 3.98EE244 pKa = 4.64LDD246 pKa = 4.37DD247 pKa = 5.59LSVSDD252 pKa = 3.97STGSNDD258 pKa = 3.74TDD260 pKa = 3.63DD261 pKa = 4.78TGDD264 pKa = 3.76SDD266 pKa = 6.2DD267 pKa = 5.19SEE269 pKa = 4.6QSTDD273 pKa = 3.7DD274 pKa = 3.49TTEE277 pKa = 4.13DD278 pKa = 3.25ATEE281 pKa = 4.43EE282 pKa = 4.24PTSDD286 pKa = 3.09TTEE289 pKa = 3.92EE290 pKa = 4.38GTNDD294 pKa = 3.27DD295 pKa = 3.97TTATTASEE303 pKa = 4.31KK304 pKa = 10.09IANEE308 pKa = 3.63IQTLQSKK315 pKa = 10.35LSNQQSIDD323 pKa = 3.54EE324 pKa = 4.67VQTQWKK330 pKa = 7.54TFASQLSQDD339 pKa = 4.04YY340 pKa = 10.58PDD342 pKa = 3.88IATQISQLTNISSALTSSEE361 pKa = 3.85QLNTFYY367 pKa = 11.55GLVLTKK373 pKa = 10.79LQTFEE378 pKa = 4.66NNSTNTNTTATSDD391 pKa = 3.43PSATLTHH398 pKa = 6.82TNTQEE403 pKa = 4.57DD404 pKa = 4.93LDD406 pKa = 3.61WLYY409 pKa = 11.28NQRR412 pKa = 11.84EE413 pKa = 4.29SHH415 pKa = 7.15DD416 pKa = 4.05NQATWW421 pKa = 2.85
MM1 pKa = 7.42YY2 pKa = 9.71YY3 pKa = 10.73SSITIISSILLATCSFVNAASTNSSYY29 pKa = 11.41PLTTINSQKK38 pKa = 10.41DD39 pKa = 3.29FCIFLPPQPGLEE51 pKa = 4.08VAVNEE56 pKa = 4.19NNGIPFCFNQDD67 pKa = 3.19AVPNATVFPTDD78 pKa = 4.72FITTAHH84 pKa = 5.95YY85 pKa = 10.57LKK87 pKa = 9.98TSTYY91 pKa = 9.02VQITGFFDD99 pKa = 3.13RR100 pKa = 11.84TKK102 pKa = 11.2YY103 pKa = 10.51DD104 pKa = 3.34LQEE107 pKa = 4.09TDD109 pKa = 4.72GGGQYY114 pKa = 10.89DD115 pKa = 3.6NHH117 pKa = 6.84AKK119 pKa = 10.07GKK121 pKa = 9.46PVGAQCKK128 pKa = 9.12GYY130 pKa = 10.66NYY132 pKa = 8.96FVNMIEE138 pKa = 4.38PDD140 pKa = 3.41LQRR143 pKa = 11.84FCIRR147 pKa = 11.84CCQDD151 pKa = 3.97KK152 pKa = 10.97DD153 pKa = 3.57DD154 pKa = 5.29CNTGRR159 pKa = 11.84SGYY162 pKa = 9.87GCLRR166 pKa = 11.84IITGDD171 pKa = 3.59YY172 pKa = 10.55TDD174 pKa = 4.86DD175 pKa = 3.59NNMIGNSTTSSGHH188 pKa = 5.7SNNHH192 pKa = 5.43INSVLAEE199 pKa = 4.21LDD201 pKa = 3.83EE202 pKa = 5.16LPAATDD208 pKa = 3.44STNATDD214 pKa = 3.64STNATDD220 pKa = 3.86STDD223 pKa = 3.39TATSADD229 pKa = 3.88TADD232 pKa = 3.79SSSTEE237 pKa = 4.19SNSALDD243 pKa = 3.98EE244 pKa = 4.64LDD246 pKa = 4.37DD247 pKa = 5.59LSVSDD252 pKa = 3.97STGSNDD258 pKa = 3.74TDD260 pKa = 3.63DD261 pKa = 4.78TGDD264 pKa = 3.76SDD266 pKa = 6.2DD267 pKa = 5.19SEE269 pKa = 4.6QSTDD273 pKa = 3.7DD274 pKa = 3.49TTEE277 pKa = 4.13DD278 pKa = 3.25ATEE281 pKa = 4.43EE282 pKa = 4.24PTSDD286 pKa = 3.09TTEE289 pKa = 3.92EE290 pKa = 4.38GTNDD294 pKa = 3.27DD295 pKa = 3.97TTATTASEE303 pKa = 4.31KK304 pKa = 10.09IANEE308 pKa = 3.63IQTLQSKK315 pKa = 10.35LSNQQSIDD323 pKa = 3.54EE324 pKa = 4.67VQTQWKK330 pKa = 7.54TFASQLSQDD339 pKa = 4.04YY340 pKa = 10.58PDD342 pKa = 3.88IATQISQLTNISSALTSSEE361 pKa = 3.85QLNTFYY367 pKa = 11.55GLVLTKK373 pKa = 10.79LQTFEE378 pKa = 4.66NNSTNTNTTATSDD391 pKa = 3.43PSATLTHH398 pKa = 6.82TNTQEE403 pKa = 4.57DD404 pKa = 4.93LDD406 pKa = 3.61WLYY409 pKa = 11.28NQRR412 pKa = 11.84EE413 pKa = 4.29SHH415 pKa = 7.15DD416 pKa = 4.05NQATWW421 pKa = 2.85
Molecular weight: 45.78 kDa
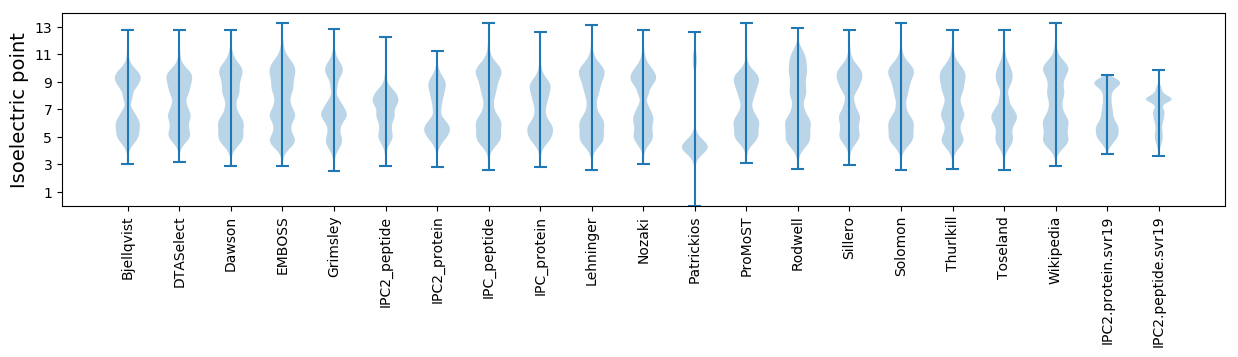
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I1C6S7|I1C6S7_RHIO9 Uncharacterized protein OS=Rhizopus delemar (strain RA 99-880 / ATCC MYA-4621 / FGSC 9543 / NRRL 43880) OX=246409 GN=RO3G_08867 PE=4 SV=1
MM1 pKa = 7.64FGQLWTTATTRR12 pKa = 11.84FASLARR18 pKa = 11.84PNTMGSSILSSSLGAFRR35 pKa = 11.84NPLTTAFNTTTQMRR49 pKa = 11.84FISRR53 pKa = 11.84GNTYY57 pKa = 10.24QPSQLVRR64 pKa = 11.84KK65 pKa = 9.19RR66 pKa = 11.84RR67 pKa = 11.84HH68 pKa = 5.1GFLARR73 pKa = 11.84LATKK77 pKa = 10.41NGRR80 pKa = 11.84HH81 pKa = 4.42TLNRR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84MKK89 pKa = 10.31GRR91 pKa = 11.84KK92 pKa = 8.59NLSHH96 pKa = 7.01
MM1 pKa = 7.64FGQLWTTATTRR12 pKa = 11.84FASLARR18 pKa = 11.84PNTMGSSILSSSLGAFRR35 pKa = 11.84NPLTTAFNTTTQMRR49 pKa = 11.84FISRR53 pKa = 11.84GNTYY57 pKa = 10.24QPSQLVRR64 pKa = 11.84KK65 pKa = 9.19RR66 pKa = 11.84RR67 pKa = 11.84HH68 pKa = 5.1GFLARR73 pKa = 11.84LATKK77 pKa = 10.41NGRR80 pKa = 11.84HH81 pKa = 4.42TLNRR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84MKK89 pKa = 10.31GRR91 pKa = 11.84KK92 pKa = 8.59NLSHH96 pKa = 7.01
Molecular weight: 10.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5840663 |
29 |
5940 |
344.2 |
39.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.339 ± 0.017 | 1.442 ± 0.008 |
5.608 ± 0.014 | 6.549 ± 0.03 |
4.018 ± 0.013 | 4.886 ± 0.018 |
2.592 ± 0.01 | 6.18 ± 0.016 |
6.793 ± 0.021 | 9.209 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.427 ± 0.007 | 5.034 ± 0.017 |
4.756 ± 0.018 | 4.484 ± 0.017 |
5.038 ± 0.014 | 8.114 ± 0.023 |
5.93 ± 0.016 | 5.939 ± 0.02 |
1.231 ± 0.007 | 3.429 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
