
Jatropha curcas (Barbados nut)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Jatropheae; Jatropha
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
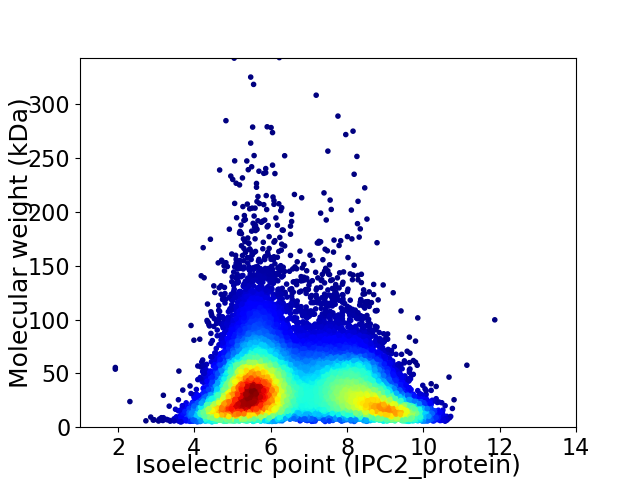
Virtual 2D-PAGE plot for 27058 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
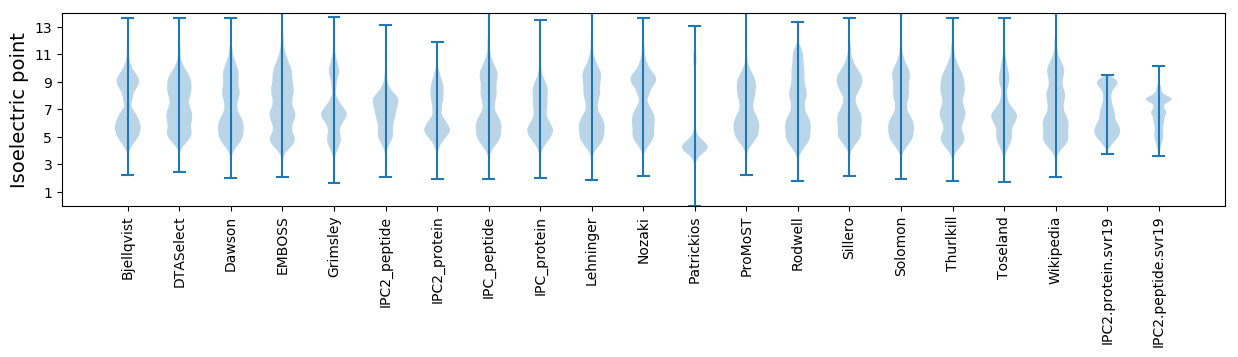
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A067JYR6|A0A067JYR6_JATCU Uncharacterized protein OS=Jatropha curcas OX=180498 GN=JCGZ_26516 PE=4 SV=1
MM1 pKa = 7.38TSSDD5 pKa = 3.36LSGEE9 pKa = 4.33DD10 pKa = 3.61FLEE13 pKa = 4.23SLGISLDD20 pKa = 4.01DD21 pKa = 4.21VDD23 pKa = 5.07LTTDD27 pKa = 3.2ADD29 pKa = 4.19TYY31 pKa = 11.66ASVTGIFAQSLEE43 pKa = 4.18NMAHH47 pKa = 6.28LAEE50 pKa = 4.6YY51 pKa = 9.94DD52 pKa = 3.13WAGAILSRR60 pKa = 11.84MYY62 pKa = 11.41DD63 pKa = 3.47DD64 pKa = 4.76MSDD67 pKa = 3.79LSRR70 pKa = 11.84GHH72 pKa = 6.61CKK74 pKa = 10.62LSGTYY79 pKa = 9.14YY80 pKa = 9.15FWEE83 pKa = 4.41VEE85 pKa = 4.34VGAMTAGLQSWIQVDD100 pKa = 3.54SHH102 pKa = 5.82FQRR105 pKa = 11.84APRR108 pKa = 11.84AISEE112 pKa = 4.28ASGPVHH118 pKa = 7.35PDD120 pKa = 2.99LANLRR125 pKa = 11.84LPYY128 pKa = 9.99SIPYY132 pKa = 8.57YY133 pKa = 10.25PPDD136 pKa = 4.18GPPALWEE143 pKa = 4.11VSLEE147 pKa = 4.34SVDD150 pKa = 5.14RR151 pKa = 11.84PALPSEE157 pKa = 5.54DD158 pKa = 3.2ITEE161 pKa = 4.31VPVGLVNQMIEE172 pKa = 4.4LMLGMQQEE180 pKa = 4.38LTAAGLIGPLTIRR193 pKa = 11.84GGGGPVADD201 pKa = 4.1SCC203 pKa = 5.47
MM1 pKa = 7.38TSSDD5 pKa = 3.36LSGEE9 pKa = 4.33DD10 pKa = 3.61FLEE13 pKa = 4.23SLGISLDD20 pKa = 4.01DD21 pKa = 4.21VDD23 pKa = 5.07LTTDD27 pKa = 3.2ADD29 pKa = 4.19TYY31 pKa = 11.66ASVTGIFAQSLEE43 pKa = 4.18NMAHH47 pKa = 6.28LAEE50 pKa = 4.6YY51 pKa = 9.94DD52 pKa = 3.13WAGAILSRR60 pKa = 11.84MYY62 pKa = 11.41DD63 pKa = 3.47DD64 pKa = 4.76MSDD67 pKa = 3.79LSRR70 pKa = 11.84GHH72 pKa = 6.61CKK74 pKa = 10.62LSGTYY79 pKa = 9.14YY80 pKa = 9.15FWEE83 pKa = 4.41VEE85 pKa = 4.34VGAMTAGLQSWIQVDD100 pKa = 3.54SHH102 pKa = 5.82FQRR105 pKa = 11.84APRR108 pKa = 11.84AISEE112 pKa = 4.28ASGPVHH118 pKa = 7.35PDD120 pKa = 2.99LANLRR125 pKa = 11.84LPYY128 pKa = 9.99SIPYY132 pKa = 8.57YY133 pKa = 10.25PPDD136 pKa = 4.18GPPALWEE143 pKa = 4.11VSLEE147 pKa = 4.34SVDD150 pKa = 5.14RR151 pKa = 11.84PALPSEE157 pKa = 5.54DD158 pKa = 3.2ITEE161 pKa = 4.31VPVGLVNQMIEE172 pKa = 4.4LMLGMQQEE180 pKa = 4.38LTAAGLIGPLTIRR193 pKa = 11.84GGGGPVADD201 pKa = 4.1SCC203 pKa = 5.47
Molecular weight: 21.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A067JDP7|A0A067JDP7_JATCU Uncharacterized protein OS=Jatropha curcas OX=180498 GN=JCGZ_03197 PE=4 SV=1
MM1 pKa = 7.7PIWYY5 pKa = 9.44LKK7 pKa = 10.51RR8 pKa = 11.84PGRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 10.37DD14 pKa = 3.1RR15 pKa = 11.84GTGPEE20 pKa = 4.02IFFGGPKK27 pKa = 9.95LPKK30 pKa = 9.85KK31 pKa = 9.97PPEE34 pKa = 3.81RR35 pKa = 11.84KK36 pKa = 8.72IRR38 pKa = 11.84RR39 pKa = 11.84SAGRR43 pKa = 11.84GGGSSRR49 pKa = 11.84PSRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84WQQPPFTAAVAAAALHH70 pKa = 6.02GGGGSSRR77 pKa = 11.84PSRR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84WQQPPFTAAVAAAALHH98 pKa = 6.02GGGGSSRR105 pKa = 11.84PSRR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84WQQPPFTAAVAAAALHH126 pKa = 6.02GGGGSSRR133 pKa = 11.84PSRR136 pKa = 11.84RR137 pKa = 11.84RR138 pKa = 11.84WQQPPFTAAVAAAALHH154 pKa = 6.02GGGGSSRR161 pKa = 11.84PSRR164 pKa = 11.84RR165 pKa = 11.84RR166 pKa = 11.84WQQPPFTAAVAAAALHH182 pKa = 6.02GGGGSSRR189 pKa = 11.84PSRR192 pKa = 11.84RR193 pKa = 11.84RR194 pKa = 11.84WQQPPFTAAVAAAALHH210 pKa = 6.02GGGGSSRR217 pKa = 11.84PSRR220 pKa = 11.84RR221 pKa = 11.84RR222 pKa = 11.84WQQPPFTAAVAAAALHH238 pKa = 6.02GGGGSSRR245 pKa = 11.84PSRR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84WQQPPFTAAVAAAALHH266 pKa = 6.02GGGGSSRR273 pKa = 11.84PSRR276 pKa = 11.84RR277 pKa = 11.84RR278 pKa = 11.84WQQPPFTAAVAAAALHH294 pKa = 6.02GGGGSSRR301 pKa = 11.84PSRR304 pKa = 11.84RR305 pKa = 11.84RR306 pKa = 11.84WQQPPFTAAVAAAALHH322 pKa = 6.02GGGGSSRR329 pKa = 11.84PSRR332 pKa = 11.84RR333 pKa = 11.84RR334 pKa = 11.84WQQPPFTAAVAAAALHH350 pKa = 6.02GGGGSSRR357 pKa = 11.84PSRR360 pKa = 11.84RR361 pKa = 11.84RR362 pKa = 11.84WQQPPFTAAVAAAALHH378 pKa = 6.02GGGGSSRR385 pKa = 11.84PSRR388 pKa = 11.84RR389 pKa = 11.84RR390 pKa = 11.84WQQPPFTAAVAAAALHH406 pKa = 6.02GGGGSSRR413 pKa = 11.84PSRR416 pKa = 11.84RR417 pKa = 11.84RR418 pKa = 11.84WQQPPFTAAVAAAALHH434 pKa = 6.02GGGGSSRR441 pKa = 11.84PSRR444 pKa = 11.84RR445 pKa = 11.84RR446 pKa = 11.84WQQPPFTAAVAAAALHH462 pKa = 6.02GGGGSSRR469 pKa = 11.84PSRR472 pKa = 11.84RR473 pKa = 11.84RR474 pKa = 11.84WQQPPFTAAVAAAALHH490 pKa = 6.02GGGGSSRR497 pKa = 11.84PSRR500 pKa = 11.84RR501 pKa = 11.84RR502 pKa = 11.84WQQPPFTAAVAAAALHH518 pKa = 6.02GGGGSSRR525 pKa = 11.84PSRR528 pKa = 11.84RR529 pKa = 11.84RR530 pKa = 11.84WQQPPFTAAVAAAALHH546 pKa = 6.02GGGGSSRR553 pKa = 11.84PSRR556 pKa = 11.84RR557 pKa = 11.84RR558 pKa = 11.84WQQPPFTAAVAAAALHH574 pKa = 6.02GGGGSSRR581 pKa = 11.84PSRR584 pKa = 11.84RR585 pKa = 11.84RR586 pKa = 11.84WQQPPFTAAVAAAALHH602 pKa = 6.02GGGGSSRR609 pKa = 11.84PSRR612 pKa = 11.84RR613 pKa = 11.84RR614 pKa = 11.84WQQPRR619 pKa = 11.84LVLQHH624 pKa = 7.02DD625 pKa = 4.16IVSGSTSGSS634 pKa = 3.26
MM1 pKa = 7.7PIWYY5 pKa = 9.44LKK7 pKa = 10.51RR8 pKa = 11.84PGRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 10.37DD14 pKa = 3.1RR15 pKa = 11.84GTGPEE20 pKa = 4.02IFFGGPKK27 pKa = 9.95LPKK30 pKa = 9.85KK31 pKa = 9.97PPEE34 pKa = 3.81RR35 pKa = 11.84KK36 pKa = 8.72IRR38 pKa = 11.84RR39 pKa = 11.84SAGRR43 pKa = 11.84GGGSSRR49 pKa = 11.84PSRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84WQQPPFTAAVAAAALHH70 pKa = 6.02GGGGSSRR77 pKa = 11.84PSRR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84WQQPPFTAAVAAAALHH98 pKa = 6.02GGGGSSRR105 pKa = 11.84PSRR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84WQQPPFTAAVAAAALHH126 pKa = 6.02GGGGSSRR133 pKa = 11.84PSRR136 pKa = 11.84RR137 pKa = 11.84RR138 pKa = 11.84WQQPPFTAAVAAAALHH154 pKa = 6.02GGGGSSRR161 pKa = 11.84PSRR164 pKa = 11.84RR165 pKa = 11.84RR166 pKa = 11.84WQQPPFTAAVAAAALHH182 pKa = 6.02GGGGSSRR189 pKa = 11.84PSRR192 pKa = 11.84RR193 pKa = 11.84RR194 pKa = 11.84WQQPPFTAAVAAAALHH210 pKa = 6.02GGGGSSRR217 pKa = 11.84PSRR220 pKa = 11.84RR221 pKa = 11.84RR222 pKa = 11.84WQQPPFTAAVAAAALHH238 pKa = 6.02GGGGSSRR245 pKa = 11.84PSRR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84WQQPPFTAAVAAAALHH266 pKa = 6.02GGGGSSRR273 pKa = 11.84PSRR276 pKa = 11.84RR277 pKa = 11.84RR278 pKa = 11.84WQQPPFTAAVAAAALHH294 pKa = 6.02GGGGSSRR301 pKa = 11.84PSRR304 pKa = 11.84RR305 pKa = 11.84RR306 pKa = 11.84WQQPPFTAAVAAAALHH322 pKa = 6.02GGGGSSRR329 pKa = 11.84PSRR332 pKa = 11.84RR333 pKa = 11.84RR334 pKa = 11.84WQQPPFTAAVAAAALHH350 pKa = 6.02GGGGSSRR357 pKa = 11.84PSRR360 pKa = 11.84RR361 pKa = 11.84RR362 pKa = 11.84WQQPPFTAAVAAAALHH378 pKa = 6.02GGGGSSRR385 pKa = 11.84PSRR388 pKa = 11.84RR389 pKa = 11.84RR390 pKa = 11.84WQQPPFTAAVAAAALHH406 pKa = 6.02GGGGSSRR413 pKa = 11.84PSRR416 pKa = 11.84RR417 pKa = 11.84RR418 pKa = 11.84WQQPPFTAAVAAAALHH434 pKa = 6.02GGGGSSRR441 pKa = 11.84PSRR444 pKa = 11.84RR445 pKa = 11.84RR446 pKa = 11.84WQQPPFTAAVAAAALHH462 pKa = 6.02GGGGSSRR469 pKa = 11.84PSRR472 pKa = 11.84RR473 pKa = 11.84RR474 pKa = 11.84WQQPPFTAAVAAAALHH490 pKa = 6.02GGGGSSRR497 pKa = 11.84PSRR500 pKa = 11.84RR501 pKa = 11.84RR502 pKa = 11.84WQQPPFTAAVAAAALHH518 pKa = 6.02GGGGSSRR525 pKa = 11.84PSRR528 pKa = 11.84RR529 pKa = 11.84RR530 pKa = 11.84WQQPPFTAAVAAAALHH546 pKa = 6.02GGGGSSRR553 pKa = 11.84PSRR556 pKa = 11.84RR557 pKa = 11.84RR558 pKa = 11.84WQQPPFTAAVAAAALHH574 pKa = 6.02GGGGSSRR581 pKa = 11.84PSRR584 pKa = 11.84RR585 pKa = 11.84RR586 pKa = 11.84WQQPPFTAAVAAAALHH602 pKa = 6.02GGGGSSRR609 pKa = 11.84PSRR612 pKa = 11.84RR613 pKa = 11.84RR614 pKa = 11.84WQQPRR619 pKa = 11.84LVLQHH624 pKa = 7.02DD625 pKa = 4.16IVSGSTSGSS634 pKa = 3.26
Molecular weight: 65.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9912802 |
50 |
5587 |
366.4 |
40.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.772 ± 0.013 | 1.808 ± 0.009 |
5.284 ± 0.011 | 6.615 ± 0.018 |
4.205 ± 0.009 | 6.525 ± 0.018 |
2.315 ± 0.007 | 5.472 ± 0.012 |
6.091 ± 0.016 | 9.673 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.427 ± 0.006 | 4.515 ± 0.01 |
5.017 ± 0.015 | 3.715 ± 0.01 |
5.374 ± 0.013 | 8.834 ± 0.017 |
4.905 ± 0.01 | 6.296 ± 0.01 |
1.305 ± 0.006 | 2.854 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
