
Hippea maritima (strain ATCC 700847 / DSM 10411 / MH2)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfurellales; Desulfurellaceae; Hippea; Hippea maritima
Average proteome isoelectric point is 7.23
Get precalculated fractions of proteins
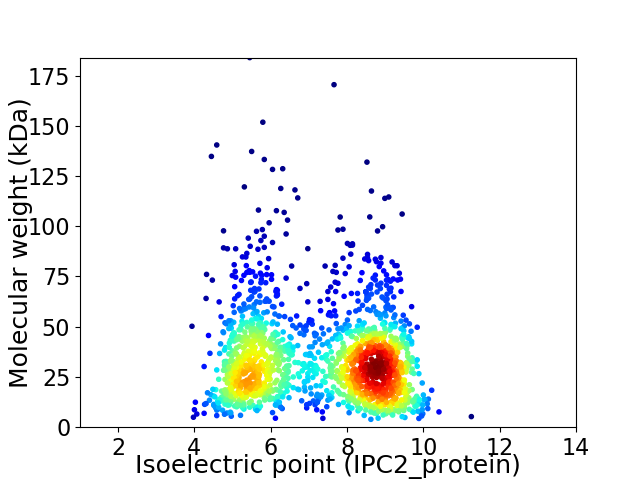
Virtual 2D-PAGE plot for 1653 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
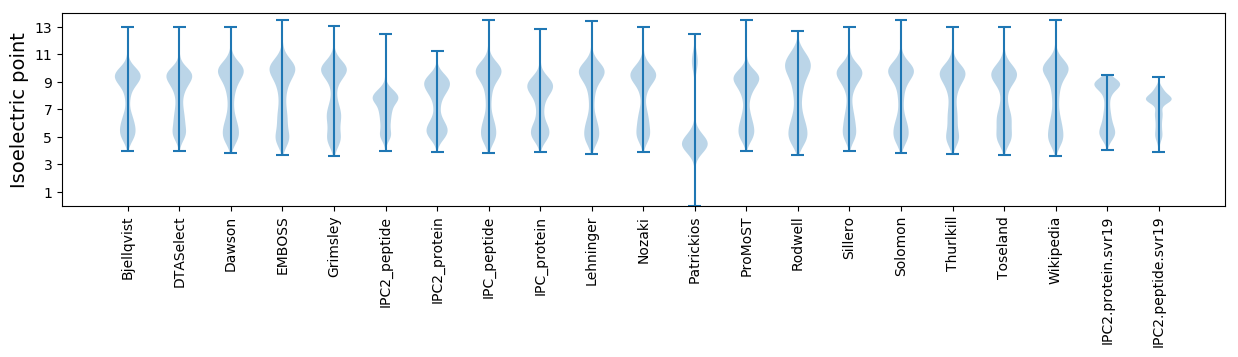
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F2LXQ7|F2LXQ7_HIPMA Uncharacterized protein OS=Hippea maritima (strain ATCC 700847 / DSM 10411 / MH2) OX=760142 GN=Hipma_1342 PE=4 SV=1
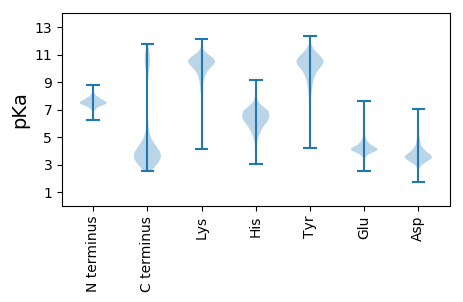
MM1 pKa = 7.62KK2 pKa = 10.03EE3 pKa = 3.7LYY5 pKa = 10.16DD6 pKa = 3.71VLFHH10 pKa = 7.19KK11 pKa = 11.13GEE13 pKa = 4.11LLVEE17 pKa = 3.96QGEE20 pKa = 4.57CEE22 pKa = 3.82KK23 pKa = 11.14AIVYY27 pKa = 7.63FLNAVKK33 pKa = 10.17EE34 pKa = 4.26NPSFEE39 pKa = 4.18DD40 pKa = 3.77AYY42 pKa = 10.3IEE44 pKa = 4.58IGYY47 pKa = 10.29CYY49 pKa = 10.5AQLDD53 pKa = 4.12NFDD56 pKa = 4.7DD57 pKa = 5.53AEE59 pKa = 4.56DD60 pKa = 3.98YY61 pKa = 10.87CNKK64 pKa = 10.38AIEE67 pKa = 4.31INPNNLEE74 pKa = 4.19AYY76 pKa = 7.54NTLAMIYY83 pKa = 9.68HH84 pKa = 6.86KK85 pKa = 10.69FGFFEE90 pKa = 5.16DD91 pKa = 4.15EE92 pKa = 3.98IEE94 pKa = 4.25ALNEE98 pKa = 3.67IIIRR102 pKa = 11.84LDD104 pKa = 3.37EE105 pKa = 5.06PDD107 pKa = 3.1ASIYY111 pKa = 10.99LNIGNAYY118 pKa = 10.04YY119 pKa = 10.56EE120 pKa = 4.24LGEE123 pKa = 3.97NDD125 pKa = 3.63RR126 pKa = 11.84AIEE129 pKa = 4.55FYY131 pKa = 11.63DD132 pKa = 3.33MAIGMEE138 pKa = 4.16PDD140 pKa = 3.24FAEE143 pKa = 5.5AYY145 pKa = 10.68ANMGNAYY152 pKa = 7.51MAKK155 pKa = 10.18DD156 pKa = 3.93EE157 pKa = 4.54YY158 pKa = 10.95IKK160 pKa = 10.53ATEE163 pKa = 4.45AYY165 pKa = 8.91KK166 pKa = 10.38QALQIDD172 pKa = 4.66PNMSDD177 pKa = 3.06VYY179 pKa = 11.03LNLGIVYY186 pKa = 10.21GEE188 pKa = 4.0LGSYY192 pKa = 10.76DD193 pKa = 3.55EE194 pKa = 4.13AVKK197 pKa = 10.92YY198 pKa = 9.94FEE200 pKa = 3.73QSIRR204 pKa = 11.84INPYY208 pKa = 9.94NPSAHH213 pKa = 6.14YY214 pKa = 9.99NLGIIWVMLNEE225 pKa = 4.29KK226 pKa = 10.06EE227 pKa = 4.23KK228 pKa = 11.1ALNEE232 pKa = 3.85YY233 pKa = 10.06EE234 pKa = 4.44RR235 pKa = 11.84LKK237 pKa = 11.17NLNKK241 pKa = 10.41DD242 pKa = 3.38LALSLVQLIDD252 pKa = 3.37KK253 pKa = 9.93EE254 pKa = 4.79SYY256 pKa = 10.54KK257 pKa = 10.35FRR259 pKa = 11.84QQ260 pKa = 3.23
MM1 pKa = 7.62KK2 pKa = 10.03EE3 pKa = 3.7LYY5 pKa = 10.16DD6 pKa = 3.71VLFHH10 pKa = 7.19KK11 pKa = 11.13GEE13 pKa = 4.11LLVEE17 pKa = 3.96QGEE20 pKa = 4.57CEE22 pKa = 3.82KK23 pKa = 11.14AIVYY27 pKa = 7.63FLNAVKK33 pKa = 10.17EE34 pKa = 4.26NPSFEE39 pKa = 4.18DD40 pKa = 3.77AYY42 pKa = 10.3IEE44 pKa = 4.58IGYY47 pKa = 10.29CYY49 pKa = 10.5AQLDD53 pKa = 4.12NFDD56 pKa = 4.7DD57 pKa = 5.53AEE59 pKa = 4.56DD60 pKa = 3.98YY61 pKa = 10.87CNKK64 pKa = 10.38AIEE67 pKa = 4.31INPNNLEE74 pKa = 4.19AYY76 pKa = 7.54NTLAMIYY83 pKa = 9.68HH84 pKa = 6.86KK85 pKa = 10.69FGFFEE90 pKa = 5.16DD91 pKa = 4.15EE92 pKa = 3.98IEE94 pKa = 4.25ALNEE98 pKa = 3.67IIIRR102 pKa = 11.84LDD104 pKa = 3.37EE105 pKa = 5.06PDD107 pKa = 3.1ASIYY111 pKa = 10.99LNIGNAYY118 pKa = 10.04YY119 pKa = 10.56EE120 pKa = 4.24LGEE123 pKa = 3.97NDD125 pKa = 3.63RR126 pKa = 11.84AIEE129 pKa = 4.55FYY131 pKa = 11.63DD132 pKa = 3.33MAIGMEE138 pKa = 4.16PDD140 pKa = 3.24FAEE143 pKa = 5.5AYY145 pKa = 10.68ANMGNAYY152 pKa = 7.51MAKK155 pKa = 10.18DD156 pKa = 3.93EE157 pKa = 4.54YY158 pKa = 10.95IKK160 pKa = 10.53ATEE163 pKa = 4.45AYY165 pKa = 8.91KK166 pKa = 10.38QALQIDD172 pKa = 4.66PNMSDD177 pKa = 3.06VYY179 pKa = 11.03LNLGIVYY186 pKa = 10.21GEE188 pKa = 4.0LGSYY192 pKa = 10.76DD193 pKa = 3.55EE194 pKa = 4.13AVKK197 pKa = 10.92YY198 pKa = 9.94FEE200 pKa = 3.73QSIRR204 pKa = 11.84INPYY208 pKa = 9.94NPSAHH213 pKa = 6.14YY214 pKa = 9.99NLGIIWVMLNEE225 pKa = 4.29KK226 pKa = 10.06EE227 pKa = 4.23KK228 pKa = 11.1ALNEE232 pKa = 3.85YY233 pKa = 10.06EE234 pKa = 4.44RR235 pKa = 11.84LKK237 pKa = 11.17NLNKK241 pKa = 10.41DD242 pKa = 3.38LALSLVQLIDD252 pKa = 3.37KK253 pKa = 9.93EE254 pKa = 4.79SYY256 pKa = 10.54KK257 pKa = 10.35FRR259 pKa = 11.84QQ260 pKa = 3.23
Molecular weight: 30.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F2LWX0|F2LWX0_HIPMA Glutaredoxin-like protein YruB-family OS=Hippea maritima (strain ATCC 700847 / DSM 10411 / MH2) OX=760142 GN=Hipma_1192 PE=4 SV=1
MM1 pKa = 7.38KK2 pKa = 9.6RR3 pKa = 11.84TFQPHH8 pKa = 5.21NKK10 pKa = 8.23PRR12 pKa = 11.84KK13 pKa = 7.28RR14 pKa = 11.84THH16 pKa = 6.07GFRR19 pKa = 11.84TRR21 pKa = 11.84MKK23 pKa = 8.41TAGGRR28 pKa = 11.84KK29 pKa = 8.24VLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.84GRR39 pKa = 11.84KK40 pKa = 8.55RR41 pKa = 11.84LSVV44 pKa = 3.2
MM1 pKa = 7.38KK2 pKa = 9.6RR3 pKa = 11.84TFQPHH8 pKa = 5.21NKK10 pKa = 8.23PRR12 pKa = 11.84KK13 pKa = 7.28RR14 pKa = 11.84THH16 pKa = 6.07GFRR19 pKa = 11.84TRR21 pKa = 11.84MKK23 pKa = 8.41TAGGRR28 pKa = 11.84KK29 pKa = 8.24VLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.84GRR39 pKa = 11.84KK40 pKa = 8.55RR41 pKa = 11.84LSVV44 pKa = 3.2
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
516646 |
35 |
1584 |
312.6 |
35.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.496 ± 0.053 | 0.941 ± 0.024 |
5.5 ± 0.048 | 6.831 ± 0.064 |
5.203 ± 0.048 | 6.476 ± 0.054 |
1.506 ± 0.021 | 8.985 ± 0.062 |
9.406 ± 0.071 | 9.511 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.298 ± 0.028 | 4.942 ± 0.052 |
3.349 ± 0.034 | 2.333 ± 0.03 |
4.123 ± 0.041 | 6.121 ± 0.053 |
4.473 ± 0.045 | 6.888 ± 0.049 |
0.704 ± 0.015 | 3.915 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
