
Candidatus Entotheonella gemina
Taxonomy: cellular organisms; Bacteria; Nitrospinae/Tectomicrobia group; Candidatus Tectomicrobia; Candidatus Entotheonella
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
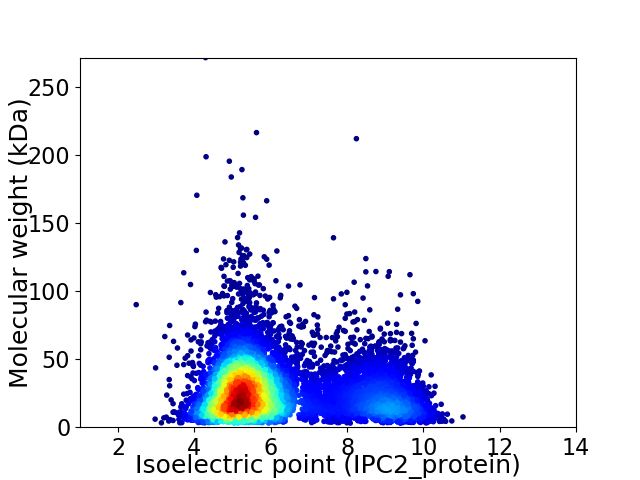
Virtual 2D-PAGE plot for 8989 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
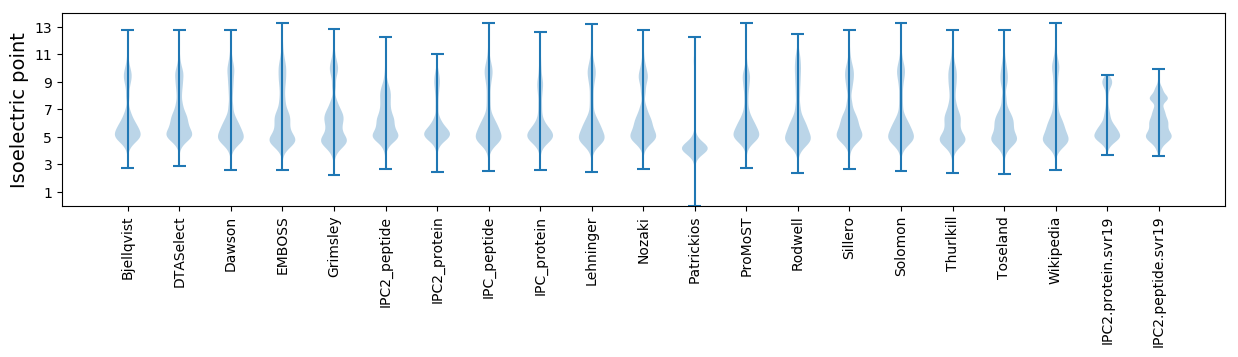
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W4M049|W4M049_9BACT Uncharacterized protein OS=Candidatus Entotheonella gemina OX=1429439 GN=ETSY2_33120 PE=4 SV=1
MM1 pKa = 7.4KK2 pKa = 10.36RR3 pKa = 11.84LSTLPASCLRR13 pKa = 11.84LMCSIIIGLVSPAWLYY29 pKa = 11.18AQLSLNEE36 pKa = 4.26NATLAQIANEE46 pKa = 4.07LEE48 pKa = 4.48GPGITIFNLSIIVGNTNQYY67 pKa = 8.69GTFTGGTIPSGAGPVVGIDD86 pKa = 3.01TGVFLTTGAVNTIAPDD102 pKa = 3.47RR103 pKa = 11.84ANITPGINDD112 pKa = 3.73NATLGPNTAGGISFSHH128 pKa = 7.29PGPPYY133 pKa = 10.4SDD135 pKa = 4.89PDD137 pKa = 3.49LTSIDD142 pKa = 3.65TQATRR147 pKa = 11.84DD148 pKa = 3.53AMIVEE153 pKa = 4.72FNVVPQQNVLKK164 pKa = 10.05IAFVFGSDD172 pKa = 3.52EE173 pKa = 4.1YY174 pKa = 10.79PEE176 pKa = 4.18YY177 pKa = 11.3VCTIFNDD184 pKa = 3.25AFGFFVTGDD193 pKa = 3.83FGSGTDD199 pKa = 3.33TTRR202 pKa = 11.84NLAVVPGTSVPIAVNTINNGSVGSAQSPGNAAPCDD237 pKa = 3.87LSNSGSFIDD246 pKa = 4.82NGDD249 pKa = 3.55GTTSSLNQNLQLDD262 pKa = 4.71GFTIPLLTQTDD273 pKa = 4.44VIPGNTYY280 pKa = 9.79RR281 pKa = 11.84VKK283 pKa = 10.77LAIADD288 pKa = 3.91ARR290 pKa = 11.84DD291 pKa = 3.86KK292 pKa = 11.17QWDD295 pKa = 3.55SAVFVNFLTSTLFNDD310 pKa = 4.6DD311 pKa = 3.1ADD313 pKa = 4.29LRR315 pKa = 11.84LSKK318 pKa = 10.66QADD321 pKa = 3.59NLAPAVGSNVTFTLTVDD338 pKa = 3.32NDD340 pKa = 4.17GPDD343 pKa = 3.24AAPGVEE349 pKa = 4.39VTDD352 pKa = 4.59LLPSGFTYY360 pKa = 10.8VSDD363 pKa = 4.06SLGGAAGGPFVDD375 pKa = 4.53YY376 pKa = 11.35NPTTGVWTLPSSVASGASATLQITATINASGDD408 pKa = 3.47YY409 pKa = 10.57TNIAEE414 pKa = 4.18ITAAQATDD422 pKa = 4.0PDD424 pKa = 4.35SEE426 pKa = 4.36PDD428 pKa = 3.59NRR430 pKa = 11.84SLNPNEE436 pKa = 5.5DD437 pKa = 3.49DD438 pKa = 3.75TASITLPPVDD448 pKa = 5.45LDD450 pKa = 3.97YY451 pKa = 11.94GDD453 pKa = 5.58APDD456 pKa = 5.06SYY458 pKa = 10.92GTDD461 pKa = 3.48STDD464 pKa = 3.03SSGEE468 pKa = 4.09GVGARR473 pKa = 11.84HH474 pKa = 7.19IINSSLYY481 pKa = 10.45LGAAVPDD488 pKa = 3.5IDD490 pKa = 4.59INGFVDD496 pKa = 6.13GIDD499 pKa = 3.79DD500 pKa = 4.27NGNGTDD506 pKa = 4.68DD507 pKa = 4.94DD508 pKa = 4.17ATGAPGNGDD517 pKa = 4.11DD518 pKa = 4.72EE519 pKa = 5.86DD520 pKa = 6.27SITSFPPLSILAAGYY535 pKa = 7.16TISGIPLHH543 pKa = 6.47NSTRR547 pKa = 11.84STAHH551 pKa = 5.33LVGWIDD557 pKa = 4.06FDD559 pKa = 4.25QSGTFDD565 pKa = 3.9ADD567 pKa = 3.15EE568 pKa = 4.48AATVSVPDD576 pKa = 3.58RR577 pKa = 11.84TSSVTLTWSSLPGIVAGVTYY597 pKa = 10.89ARR599 pKa = 11.84FRR601 pKa = 11.84LTTDD605 pKa = 3.22TSIATGTAATSVPTGTAIDD624 pKa = 4.56GEE626 pKa = 4.55VEE628 pKa = 4.26DD629 pKa = 4.17YY630 pKa = 11.15QLIIGGLTVSGTVYY644 pKa = 11.13NDD646 pKa = 3.42ANHH649 pKa = 6.04NRR651 pKa = 11.84QLDD654 pKa = 3.7SGEE657 pKa = 4.28TGTGLTLFAKK667 pKa = 9.53LIPASTPAGPALSAVPVDD685 pKa = 4.16AGSGAYY691 pKa = 10.17AFTSVNPLTYY701 pKa = 10.08RR702 pKa = 11.84VIIDD706 pKa = 4.13DD707 pKa = 4.04NATLSDD713 pKa = 3.85VTPTLPAGWIGTEE726 pKa = 3.58IPGQIRR732 pKa = 11.84PAVTVVNASVPNQNFGLFNGSQLSGTVFIDD762 pKa = 3.1NGADD766 pKa = 3.05RR767 pKa = 11.84GTPNNGLQDD776 pKa = 3.6GGEE779 pKa = 4.19TGLGSVTVTATDD791 pKa = 3.91GASTVYY797 pKa = 9.74DD798 pKa = 3.86TTTTAADD805 pKa = 3.72GTYY808 pKa = 9.93TLYY811 pKa = 11.07VPAAATTVAVTQTNLSAYY829 pKa = 9.33LSTGGSPGTTGGTYY843 pKa = 10.46DD844 pKa = 4.19RR845 pKa = 11.84NPDD848 pKa = 3.87TVSLTATAGTSYY860 pKa = 10.85SGVDD864 pKa = 4.38FGDD867 pKa = 3.58VPNNTFVPDD876 pKa = 4.12GQQNGLPGTVLWYY889 pKa = 9.5PHH891 pKa = 6.41TFTAGSAGTVTFSTSGSSSPAIPGWSEE918 pKa = 3.91TLYY921 pKa = 10.91QDD923 pKa = 3.99NNCNSIIDD931 pKa = 3.86NGEE934 pKa = 3.86GQIGSIALSAGDD946 pKa = 3.57QVCILMRR953 pKa = 11.84EE954 pKa = 5.06AIPHH958 pKa = 6.61AAPDD962 pKa = 3.57GATRR966 pKa = 11.84QVTVSASFSYY976 pKa = 11.34DD977 pKa = 2.99NASPSLSHH985 pKa = 5.69VLQRR989 pKa = 11.84IDD991 pKa = 3.48LTTVGDD997 pKa = 3.85STSAGLHH1004 pKa = 5.82LLKK1007 pKa = 10.85AVDD1010 pKa = 3.78KK1011 pKa = 8.98TQAAPGEE1018 pKa = 4.33VLTYY1022 pKa = 10.18TITYY1026 pKa = 9.37SNRR1029 pKa = 11.84SNEE1032 pKa = 4.12SLSNIVIHH1040 pKa = 7.22DD1041 pKa = 4.24LTPAFTVFQSATCDD1055 pKa = 3.28LPLPLDD1061 pKa = 3.37ISGCNVTQQPVINGTGAIEE1080 pKa = 3.99WTLGGTLRR1088 pKa = 11.84PGHH1091 pKa = 6.06QGTVTFSIQIRR1102 pKa = 11.84QQ1103 pKa = 3.21
MM1 pKa = 7.4KK2 pKa = 10.36RR3 pKa = 11.84LSTLPASCLRR13 pKa = 11.84LMCSIIIGLVSPAWLYY29 pKa = 11.18AQLSLNEE36 pKa = 4.26NATLAQIANEE46 pKa = 4.07LEE48 pKa = 4.48GPGITIFNLSIIVGNTNQYY67 pKa = 8.69GTFTGGTIPSGAGPVVGIDD86 pKa = 3.01TGVFLTTGAVNTIAPDD102 pKa = 3.47RR103 pKa = 11.84ANITPGINDD112 pKa = 3.73NATLGPNTAGGISFSHH128 pKa = 7.29PGPPYY133 pKa = 10.4SDD135 pKa = 4.89PDD137 pKa = 3.49LTSIDD142 pKa = 3.65TQATRR147 pKa = 11.84DD148 pKa = 3.53AMIVEE153 pKa = 4.72FNVVPQQNVLKK164 pKa = 10.05IAFVFGSDD172 pKa = 3.52EE173 pKa = 4.1YY174 pKa = 10.79PEE176 pKa = 4.18YY177 pKa = 11.3VCTIFNDD184 pKa = 3.25AFGFFVTGDD193 pKa = 3.83FGSGTDD199 pKa = 3.33TTRR202 pKa = 11.84NLAVVPGTSVPIAVNTINNGSVGSAQSPGNAAPCDD237 pKa = 3.87LSNSGSFIDD246 pKa = 4.82NGDD249 pKa = 3.55GTTSSLNQNLQLDD262 pKa = 4.71GFTIPLLTQTDD273 pKa = 4.44VIPGNTYY280 pKa = 9.79RR281 pKa = 11.84VKK283 pKa = 10.77LAIADD288 pKa = 3.91ARR290 pKa = 11.84DD291 pKa = 3.86KK292 pKa = 11.17QWDD295 pKa = 3.55SAVFVNFLTSTLFNDD310 pKa = 4.6DD311 pKa = 3.1ADD313 pKa = 4.29LRR315 pKa = 11.84LSKK318 pKa = 10.66QADD321 pKa = 3.59NLAPAVGSNVTFTLTVDD338 pKa = 3.32NDD340 pKa = 4.17GPDD343 pKa = 3.24AAPGVEE349 pKa = 4.39VTDD352 pKa = 4.59LLPSGFTYY360 pKa = 10.8VSDD363 pKa = 4.06SLGGAAGGPFVDD375 pKa = 4.53YY376 pKa = 11.35NPTTGVWTLPSSVASGASATLQITATINASGDD408 pKa = 3.47YY409 pKa = 10.57TNIAEE414 pKa = 4.18ITAAQATDD422 pKa = 4.0PDD424 pKa = 4.35SEE426 pKa = 4.36PDD428 pKa = 3.59NRR430 pKa = 11.84SLNPNEE436 pKa = 5.5DD437 pKa = 3.49DD438 pKa = 3.75TASITLPPVDD448 pKa = 5.45LDD450 pKa = 3.97YY451 pKa = 11.94GDD453 pKa = 5.58APDD456 pKa = 5.06SYY458 pKa = 10.92GTDD461 pKa = 3.48STDD464 pKa = 3.03SSGEE468 pKa = 4.09GVGARR473 pKa = 11.84HH474 pKa = 7.19IINSSLYY481 pKa = 10.45LGAAVPDD488 pKa = 3.5IDD490 pKa = 4.59INGFVDD496 pKa = 6.13GIDD499 pKa = 3.79DD500 pKa = 4.27NGNGTDD506 pKa = 4.68DD507 pKa = 4.94DD508 pKa = 4.17ATGAPGNGDD517 pKa = 4.11DD518 pKa = 4.72EE519 pKa = 5.86DD520 pKa = 6.27SITSFPPLSILAAGYY535 pKa = 7.16TISGIPLHH543 pKa = 6.47NSTRR547 pKa = 11.84STAHH551 pKa = 5.33LVGWIDD557 pKa = 4.06FDD559 pKa = 4.25QSGTFDD565 pKa = 3.9ADD567 pKa = 3.15EE568 pKa = 4.48AATVSVPDD576 pKa = 3.58RR577 pKa = 11.84TSSVTLTWSSLPGIVAGVTYY597 pKa = 10.89ARR599 pKa = 11.84FRR601 pKa = 11.84LTTDD605 pKa = 3.22TSIATGTAATSVPTGTAIDD624 pKa = 4.56GEE626 pKa = 4.55VEE628 pKa = 4.26DD629 pKa = 4.17YY630 pKa = 11.15QLIIGGLTVSGTVYY644 pKa = 11.13NDD646 pKa = 3.42ANHH649 pKa = 6.04NRR651 pKa = 11.84QLDD654 pKa = 3.7SGEE657 pKa = 4.28TGTGLTLFAKK667 pKa = 9.53LIPASTPAGPALSAVPVDD685 pKa = 4.16AGSGAYY691 pKa = 10.17AFTSVNPLTYY701 pKa = 10.08RR702 pKa = 11.84VIIDD706 pKa = 4.13DD707 pKa = 4.04NATLSDD713 pKa = 3.85VTPTLPAGWIGTEE726 pKa = 3.58IPGQIRR732 pKa = 11.84PAVTVVNASVPNQNFGLFNGSQLSGTVFIDD762 pKa = 3.1NGADD766 pKa = 3.05RR767 pKa = 11.84GTPNNGLQDD776 pKa = 3.6GGEE779 pKa = 4.19TGLGSVTVTATDD791 pKa = 3.91GASTVYY797 pKa = 9.74DD798 pKa = 3.86TTTTAADD805 pKa = 3.72GTYY808 pKa = 9.93TLYY811 pKa = 11.07VPAAATTVAVTQTNLSAYY829 pKa = 9.33LSTGGSPGTTGGTYY843 pKa = 10.46DD844 pKa = 4.19RR845 pKa = 11.84NPDD848 pKa = 3.87TVSLTATAGTSYY860 pKa = 10.85SGVDD864 pKa = 4.38FGDD867 pKa = 3.58VPNNTFVPDD876 pKa = 4.12GQQNGLPGTVLWYY889 pKa = 9.5PHH891 pKa = 6.41TFTAGSAGTVTFSTSGSSSPAIPGWSEE918 pKa = 3.91TLYY921 pKa = 10.91QDD923 pKa = 3.99NNCNSIIDD931 pKa = 3.86NGEE934 pKa = 3.86GQIGSIALSAGDD946 pKa = 3.57QVCILMRR953 pKa = 11.84EE954 pKa = 5.06AIPHH958 pKa = 6.61AAPDD962 pKa = 3.57GATRR966 pKa = 11.84QVTVSASFSYY976 pKa = 11.34DD977 pKa = 2.99NASPSLSHH985 pKa = 5.69VLQRR989 pKa = 11.84IDD991 pKa = 3.48LTTVGDD997 pKa = 3.85STSAGLHH1004 pKa = 5.82LLKK1007 pKa = 10.85AVDD1010 pKa = 3.78KK1011 pKa = 8.98TQAAPGEE1018 pKa = 4.33VLTYY1022 pKa = 10.18TITYY1026 pKa = 9.37SNRR1029 pKa = 11.84SNEE1032 pKa = 4.12SLSNIVIHH1040 pKa = 7.22DD1041 pKa = 4.24LTPAFTVFQSATCDD1055 pKa = 3.28LPLPLDD1061 pKa = 3.37ISGCNVTQQPVINGTGAIEE1080 pKa = 3.99WTLGGTLRR1088 pKa = 11.84PGHH1091 pKa = 6.06QGTVTFSIQIRR1102 pKa = 11.84QQ1103 pKa = 3.21
Molecular weight: 113.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W4LQR2|W4LQR2_9BACT Translation initiation factor IF-3 OS=Candidatus Entotheonella gemina OX=1429439 GN=infC PE=3 SV=1
MM1 pKa = 7.89PKK3 pKa = 9.71MKK5 pKa = 9.95TNRR8 pKa = 11.84SAAKK12 pKa = 10.05RR13 pKa = 11.84FRR15 pKa = 11.84LTARR19 pKa = 11.84GKK21 pKa = 9.33VRR23 pKa = 11.84RR24 pKa = 11.84NQAFTRR30 pKa = 11.84HH31 pKa = 5.85ILTKK35 pKa = 9.37KK36 pKa = 6.62TRR38 pKa = 11.84KK39 pKa = 9.41RR40 pKa = 11.84KK41 pKa = 8.77RR42 pKa = 11.84QLRR45 pKa = 11.84PTALVAAADD54 pKa = 3.61APRR57 pKa = 11.84IKK59 pKa = 10.42RR60 pKa = 11.84IITRR64 pKa = 3.66
MM1 pKa = 7.89PKK3 pKa = 9.71MKK5 pKa = 9.95TNRR8 pKa = 11.84SAAKK12 pKa = 10.05RR13 pKa = 11.84FRR15 pKa = 11.84LTARR19 pKa = 11.84GKK21 pKa = 9.33VRR23 pKa = 11.84RR24 pKa = 11.84NQAFTRR30 pKa = 11.84HH31 pKa = 5.85ILTKK35 pKa = 9.37KK36 pKa = 6.62TRR38 pKa = 11.84KK39 pKa = 9.41RR40 pKa = 11.84KK41 pKa = 8.77RR42 pKa = 11.84QLRR45 pKa = 11.84PTALVAAADD54 pKa = 3.61APRR57 pKa = 11.84IKK59 pKa = 10.42RR60 pKa = 11.84IITRR64 pKa = 3.66
Molecular weight: 7.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2328076 |
29 |
2567 |
259.0 |
28.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.011 ± 0.031 | 1.08 ± 0.009 |
5.677 ± 0.023 | 6.135 ± 0.026 |
3.67 ± 0.017 | 7.572 ± 0.026 |
2.748 ± 0.015 | 5.319 ± 0.022 |
2.981 ± 0.022 | 10.363 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.593 ± 0.015 | 2.786 ± 0.015 |
5.296 ± 0.019 | 4.501 ± 0.023 |
6.83 ± 0.021 | 5.334 ± 0.017 |
5.541 ± 0.019 | 7.146 ± 0.022 |
1.508 ± 0.012 | 2.911 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
