
Chlorella variabilis (Green alga)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; core chlorophytes; Trebouxiophyceae; Chlorellales; Chlorellaceae; Chlorella clade; Chlorella
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
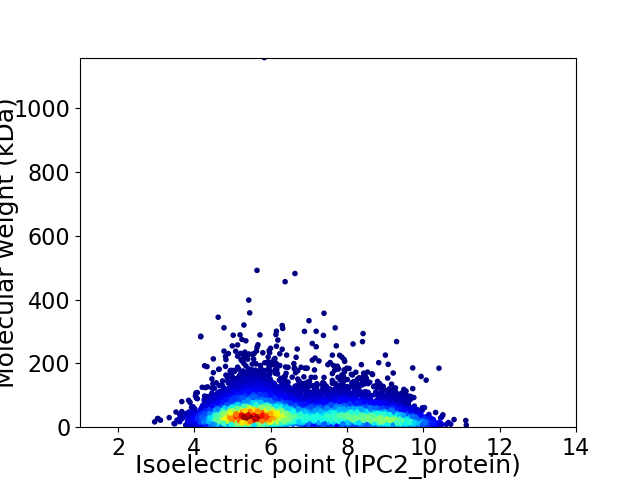
Virtual 2D-PAGE plot for 9831 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E1ZK52|E1ZK52_CHLVA Expressed protein OS=Chlorella variabilis OX=554065 GN=CHLNCDRAFT_136332 PE=4 SV=1
MM1 pKa = 7.42ARR3 pKa = 11.84HH4 pKa = 6.04PAGLAAPAALLLILAAGAWTARR26 pKa = 11.84AASDD30 pKa = 3.98CPYY33 pKa = 9.6MNTSEE38 pKa = 4.5PVPATWNFQLSGPLVDD54 pKa = 6.5DD55 pKa = 4.76FMQPDD60 pKa = 3.63MMGKK64 pKa = 9.32VPGWLTTAFEE74 pKa = 4.36ASDD77 pKa = 4.68LFTIDD82 pKa = 3.65PSLTFGTYY90 pKa = 8.31EE91 pKa = 3.77YY92 pKa = 10.45RR93 pKa = 11.84DD94 pKa = 3.66QAEE97 pKa = 4.29VTSPYY102 pKa = 10.85ASYY105 pKa = 11.02GAIFNLTSPASEE117 pKa = 4.03IFGQDD122 pKa = 3.29VIDD125 pKa = 3.97EE126 pKa = 5.4LIPNDD131 pKa = 3.68EE132 pKa = 3.87IGVFKK137 pKa = 10.78RR138 pKa = 11.84QFFHH142 pKa = 7.4VSGTGSHH149 pKa = 5.59VSAVLSFVDD158 pKa = 4.74YY159 pKa = 10.14EE160 pKa = 4.29ALNITEE166 pKa = 4.33PTTLSSNSSVDD177 pKa = 3.65FAFYY181 pKa = 11.32VLGCTVQIPQAFTFSTTPPPPGLSITDD208 pKa = 3.81YY209 pKa = 11.32EE210 pKa = 4.68DD211 pKa = 3.3VAEE214 pKa = 4.77LLPSFATWVPSPDD227 pKa = 3.13SVANTTSPGFAFYY240 pKa = 10.22QYY242 pKa = 9.98WPLVVKK248 pKa = 10.84ASGEE252 pKa = 4.1PIPVFVRR259 pKa = 11.84LVTTLNSDD267 pKa = 3.46SLEE270 pKa = 4.08AAKK273 pKa = 10.63QGVNSSGTGFDD284 pKa = 3.85IYY286 pKa = 10.49WLAGDD291 pKa = 3.73EE292 pKa = 4.57VVAEE296 pKa = 4.15EE297 pKa = 4.67AGLEE301 pKa = 3.89ASLEE305 pKa = 4.19EE306 pKa = 4.24VHH308 pKa = 7.1ANLAAVLNRR317 pKa = 11.84LNDD320 pKa = 3.69TMNGGGDD327 pKa = 3.84SGPAPAPAPQPASGSAIGAAAGTPMLMALALATLAFLAA365 pKa = 4.77
MM1 pKa = 7.42ARR3 pKa = 11.84HH4 pKa = 6.04PAGLAAPAALLLILAAGAWTARR26 pKa = 11.84AASDD30 pKa = 3.98CPYY33 pKa = 9.6MNTSEE38 pKa = 4.5PVPATWNFQLSGPLVDD54 pKa = 6.5DD55 pKa = 4.76FMQPDD60 pKa = 3.63MMGKK64 pKa = 9.32VPGWLTTAFEE74 pKa = 4.36ASDD77 pKa = 4.68LFTIDD82 pKa = 3.65PSLTFGTYY90 pKa = 8.31EE91 pKa = 3.77YY92 pKa = 10.45RR93 pKa = 11.84DD94 pKa = 3.66QAEE97 pKa = 4.29VTSPYY102 pKa = 10.85ASYY105 pKa = 11.02GAIFNLTSPASEE117 pKa = 4.03IFGQDD122 pKa = 3.29VIDD125 pKa = 3.97EE126 pKa = 5.4LIPNDD131 pKa = 3.68EE132 pKa = 3.87IGVFKK137 pKa = 10.78RR138 pKa = 11.84QFFHH142 pKa = 7.4VSGTGSHH149 pKa = 5.59VSAVLSFVDD158 pKa = 4.74YY159 pKa = 10.14EE160 pKa = 4.29ALNITEE166 pKa = 4.33PTTLSSNSSVDD177 pKa = 3.65FAFYY181 pKa = 11.32VLGCTVQIPQAFTFSTTPPPPGLSITDD208 pKa = 3.81YY209 pKa = 11.32EE210 pKa = 4.68DD211 pKa = 3.3VAEE214 pKa = 4.77LLPSFATWVPSPDD227 pKa = 3.13SVANTTSPGFAFYY240 pKa = 10.22QYY242 pKa = 9.98WPLVVKK248 pKa = 10.84ASGEE252 pKa = 4.1PIPVFVRR259 pKa = 11.84LVTTLNSDD267 pKa = 3.46SLEE270 pKa = 4.08AAKK273 pKa = 10.63QGVNSSGTGFDD284 pKa = 3.85IYY286 pKa = 10.49WLAGDD291 pKa = 3.73EE292 pKa = 4.57VVAEE296 pKa = 4.15EE297 pKa = 4.67AGLEE301 pKa = 3.89ASLEE305 pKa = 4.19EE306 pKa = 4.24VHH308 pKa = 7.1ANLAAVLNRR317 pKa = 11.84LNDD320 pKa = 3.69TMNGGGDD327 pKa = 3.84SGPAPAPAPQPASGSAIGAAAGTPMLMALALATLAFLAA365 pKa = 4.77
Molecular weight: 38.41 kDa
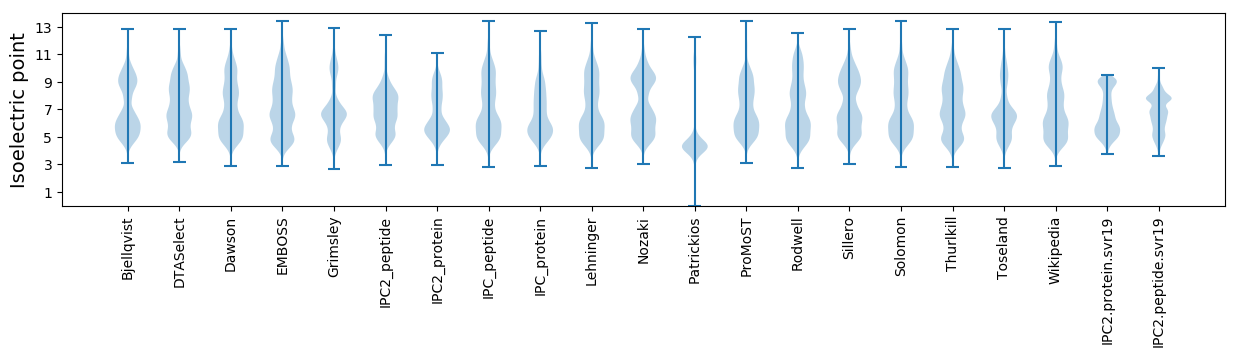
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E1Z1V6|E1Z1V6_CHLVA J domain-containing protein OS=Chlorella variabilis OX=554065 GN=CHLNCDRAFT_18104 PE=4 SV=1
MM1 pKa = 7.24CRR3 pKa = 11.84ARR5 pKa = 11.84RR6 pKa = 11.84GPQTQTWPRR15 pKa = 11.84STSRR19 pKa = 11.84AAAWAQPGRR28 pKa = 11.84QRR30 pKa = 11.84PRR32 pKa = 11.84WRR34 pKa = 11.84RR35 pKa = 11.84LRR37 pKa = 11.84RR38 pKa = 11.84GAGTRR43 pKa = 11.84PRR45 pKa = 11.84APPPPPTRR53 pKa = 11.84QRR55 pKa = 11.84APRR58 pKa = 11.84RR59 pKa = 11.84APPRR63 pKa = 11.84PRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84GAGSRR73 pKa = 11.84RR74 pKa = 11.84ASAPRR79 pKa = 11.84RR80 pKa = 11.84PSLLLRR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84GQRR94 pKa = 11.84ASPRR98 pKa = 11.84RR99 pKa = 11.84KK100 pKa = 9.51AGSSSRR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84HH109 pKa = 5.38AAPSRR114 pKa = 11.84CQAGTPSPRR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84LAAAQPRR132 pKa = 11.84PAAATPAPARR142 pKa = 11.84ALATPPCLEE151 pKa = 3.57WRR153 pKa = 11.84RR154 pKa = 11.84WCPACPARR162 pKa = 11.84RR163 pKa = 11.84RR164 pKa = 11.84RR165 pKa = 11.84RR166 pKa = 11.84RR167 pKa = 11.84GRR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 11.84RR172 pKa = 11.84ASWRR176 pKa = 11.84RR177 pKa = 11.84PGRR180 pKa = 11.84RR181 pKa = 3.17
MM1 pKa = 7.24CRR3 pKa = 11.84ARR5 pKa = 11.84RR6 pKa = 11.84GPQTQTWPRR15 pKa = 11.84STSRR19 pKa = 11.84AAAWAQPGRR28 pKa = 11.84QRR30 pKa = 11.84PRR32 pKa = 11.84WRR34 pKa = 11.84RR35 pKa = 11.84LRR37 pKa = 11.84RR38 pKa = 11.84GAGTRR43 pKa = 11.84PRR45 pKa = 11.84APPPPPTRR53 pKa = 11.84QRR55 pKa = 11.84APRR58 pKa = 11.84RR59 pKa = 11.84APPRR63 pKa = 11.84PRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84GAGSRR73 pKa = 11.84RR74 pKa = 11.84ASAPRR79 pKa = 11.84RR80 pKa = 11.84PSLLLRR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84GQRR94 pKa = 11.84ASPRR98 pKa = 11.84RR99 pKa = 11.84KK100 pKa = 9.51AGSSSRR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84HH109 pKa = 5.38AAPSRR114 pKa = 11.84CQAGTPSPRR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84LAAAQPRR132 pKa = 11.84PAAATPAPARR142 pKa = 11.84ALATPPCLEE151 pKa = 3.57WRR153 pKa = 11.84RR154 pKa = 11.84WCPACPARR162 pKa = 11.84RR163 pKa = 11.84RR164 pKa = 11.84RR165 pKa = 11.84RR166 pKa = 11.84RR167 pKa = 11.84GRR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 11.84RR172 pKa = 11.84ASWRR176 pKa = 11.84RR177 pKa = 11.84PGRR180 pKa = 11.84RR181 pKa = 3.17
Molecular weight: 20.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4464018 |
31 |
11464 |
454.1 |
48.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.121 ± 0.062 | 1.736 ± 0.016 |
4.409 ± 0.017 | 5.908 ± 0.028 |
2.696 ± 0.017 | 9.222 ± 0.029 |
2.223 ± 0.01 | 2.54 ± 0.018 |
3.09 ± 0.025 | 9.998 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.984 ± 0.01 | 1.949 ± 0.015 |
6.406 ± 0.029 | 5.563 ± 0.035 |
6.663 ± 0.022 | 6.449 ± 0.024 |
4.243 ± 0.02 | 6.39 ± 0.02 |
1.429 ± 0.01 | 1.982 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
