
Desulfurella amilsii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfurellales; Desulfurellaceae; Desulfurella
Average proteome isoelectric point is 7.16
Get precalculated fractions of proteins
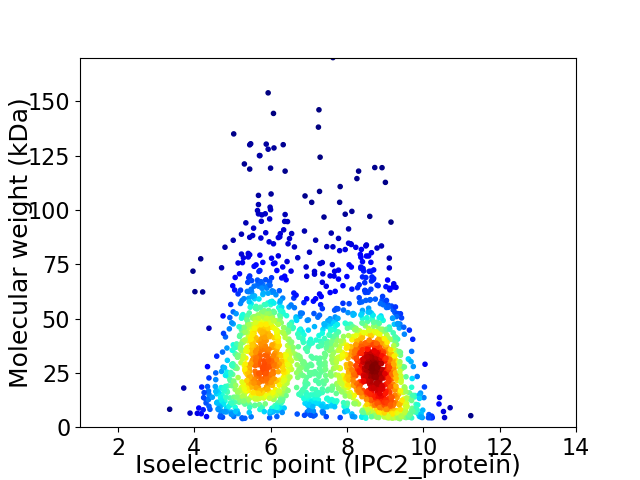
Virtual 2D-PAGE plot for 2019 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X4XUT4|A0A1X4XUT4_9DELT Uncharacterized protein OS=Desulfurella amilsii OX=1562698 GN=DESAMIL20_843 PE=4 SV=1
MM1 pKa = 7.09MRR3 pKa = 11.84SLWSGVSGLQTSQDD17 pKa = 3.36FMDD20 pKa = 3.58VVGNNVANVNTVGYY34 pKa = 9.38KK35 pKa = 10.15ASQINFSDD43 pKa = 3.73VLAQTLSGATGPQGALGGMNPMQIGLGTQVASTSKK78 pKa = 10.5NFSQGSLQTTNVTTNLAIQGNGFFIVSPDD107 pKa = 3.18NGNTYY112 pKa = 10.66SYY114 pKa = 10.64TRR116 pKa = 11.84AGDD119 pKa = 3.42FTFDD123 pKa = 3.24AKK125 pKa = 11.34GNLVTPTGDD134 pKa = 3.55VVQGWLANNTTHH146 pKa = 8.07LINTASTIQGINIPQNMTVPAGVTKK171 pKa = 10.4NVSMSGNLDD180 pKa = 3.36GGQTVQTSEE189 pKa = 4.13LTAITPSLEE198 pKa = 4.01NSTGAINMNSIYY210 pKa = 10.61NSNGNLIGVGQTTATAAVGAGSDD233 pKa = 4.03TVSGLYY239 pKa = 10.12DD240 pKa = 3.25INGNQIGSNWTVTVGGTTLTPTPATLSDD268 pKa = 4.26LANALAPSGTTATITPGGEE287 pKa = 3.81IQISSTGGTTLPTITSSNPVLNGILSNLSGKK318 pKa = 7.85TIPAGGTITSLPFFSTPGDD337 pKa = 3.68TVAMSFDD344 pKa = 3.1GGANYY349 pKa = 7.76NTYY352 pKa = 10.15IYY354 pKa = 10.95GSGTGGGGTRR364 pKa = 11.84NSTTGIMSGNVQYY377 pKa = 8.36FTTLDD382 pKa = 3.77DD383 pKa = 4.55LKK385 pKa = 11.43SDD387 pKa = 3.37INTDD391 pKa = 3.13INDD394 pKa = 3.68NSATPPINNATVTLNSSGEE413 pKa = 4.06MQIVPTTGSNLSSIGPVYY431 pKa = 10.99SNNSNLATILGTLSGQVQQTGSTSQPFMADD461 pKa = 3.04TYY463 pKa = 8.33STSVAVYY470 pKa = 10.08DD471 pKa = 4.21SLGNKK476 pKa = 9.59HH477 pKa = 6.86DD478 pKa = 3.63VTFNFAKK485 pKa = 10.13TNYY488 pKa = 9.58NPSTNHH494 pKa = 5.71TQWEE498 pKa = 4.95WYY500 pKa = 7.91ATAPSSGSSTPTLTNASGQITFDD523 pKa = 3.45STGKK527 pKa = 10.48LVGLTPPLAITANWNNGTSQQVVNLHH553 pKa = 6.42FGDD556 pKa = 4.73LNTFDD561 pKa = 5.51GLTQFSLPSQTSSLTQDD578 pKa = 3.54GYY580 pKa = 11.89AGGSLQNIIVNQNGVISGMFSNGKK604 pKa = 9.72SYY606 pKa = 11.6ALGQVSLATFNNNDD620 pKa = 3.31GLLSEE625 pKa = 4.74GNSLFEE631 pKa = 4.01ATANSGTPIVTTAGTGTAGTVIPSEE656 pKa = 4.24LEE658 pKa = 3.75EE659 pKa = 4.93SNVDD663 pKa = 3.88LGTEE667 pKa = 4.4FTNMIIAEE675 pKa = 4.11RR676 pKa = 11.84AYY678 pKa = 10.04QANARR683 pKa = 11.84TLSTSDD689 pKa = 3.18TMLQSLLNIQQ699 pKa = 3.52
MM1 pKa = 7.09MRR3 pKa = 11.84SLWSGVSGLQTSQDD17 pKa = 3.36FMDD20 pKa = 3.58VVGNNVANVNTVGYY34 pKa = 9.38KK35 pKa = 10.15ASQINFSDD43 pKa = 3.73VLAQTLSGATGPQGALGGMNPMQIGLGTQVASTSKK78 pKa = 10.5NFSQGSLQTTNVTTNLAIQGNGFFIVSPDD107 pKa = 3.18NGNTYY112 pKa = 10.66SYY114 pKa = 10.64TRR116 pKa = 11.84AGDD119 pKa = 3.42FTFDD123 pKa = 3.24AKK125 pKa = 11.34GNLVTPTGDD134 pKa = 3.55VVQGWLANNTTHH146 pKa = 8.07LINTASTIQGINIPQNMTVPAGVTKK171 pKa = 10.4NVSMSGNLDD180 pKa = 3.36GGQTVQTSEE189 pKa = 4.13LTAITPSLEE198 pKa = 4.01NSTGAINMNSIYY210 pKa = 10.61NSNGNLIGVGQTTATAAVGAGSDD233 pKa = 4.03TVSGLYY239 pKa = 10.12DD240 pKa = 3.25INGNQIGSNWTVTVGGTTLTPTPATLSDD268 pKa = 4.26LANALAPSGTTATITPGGEE287 pKa = 3.81IQISSTGGTTLPTITSSNPVLNGILSNLSGKK318 pKa = 7.85TIPAGGTITSLPFFSTPGDD337 pKa = 3.68TVAMSFDD344 pKa = 3.1GGANYY349 pKa = 7.76NTYY352 pKa = 10.15IYY354 pKa = 10.95GSGTGGGGTRR364 pKa = 11.84NSTTGIMSGNVQYY377 pKa = 8.36FTTLDD382 pKa = 3.77DD383 pKa = 4.55LKK385 pKa = 11.43SDD387 pKa = 3.37INTDD391 pKa = 3.13INDD394 pKa = 3.68NSATPPINNATVTLNSSGEE413 pKa = 4.06MQIVPTTGSNLSSIGPVYY431 pKa = 10.99SNNSNLATILGTLSGQVQQTGSTSQPFMADD461 pKa = 3.04TYY463 pKa = 8.33STSVAVYY470 pKa = 10.08DD471 pKa = 4.21SLGNKK476 pKa = 9.59HH477 pKa = 6.86DD478 pKa = 3.63VTFNFAKK485 pKa = 10.13TNYY488 pKa = 9.58NPSTNHH494 pKa = 5.71TQWEE498 pKa = 4.95WYY500 pKa = 7.91ATAPSSGSSTPTLTNASGQITFDD523 pKa = 3.45STGKK527 pKa = 10.48LVGLTPPLAITANWNNGTSQQVVNLHH553 pKa = 6.42FGDD556 pKa = 4.73LNTFDD561 pKa = 5.51GLTQFSLPSQTSSLTQDD578 pKa = 3.54GYY580 pKa = 11.89AGGSLQNIIVNQNGVISGMFSNGKK604 pKa = 9.72SYY606 pKa = 11.6ALGQVSLATFNNNDD620 pKa = 3.31GLLSEE625 pKa = 4.74GNSLFEE631 pKa = 4.01ATANSGTPIVTTAGTGTAGTVIPSEE656 pKa = 4.24LEE658 pKa = 3.75EE659 pKa = 4.93SNVDD663 pKa = 3.88LGTEE667 pKa = 4.4FTNMIIAEE675 pKa = 4.11RR676 pKa = 11.84AYY678 pKa = 10.04QANARR683 pKa = 11.84TLSTSDD689 pKa = 3.18TMLQSLLNIQQ699 pKa = 3.52
Molecular weight: 71.79 kDa
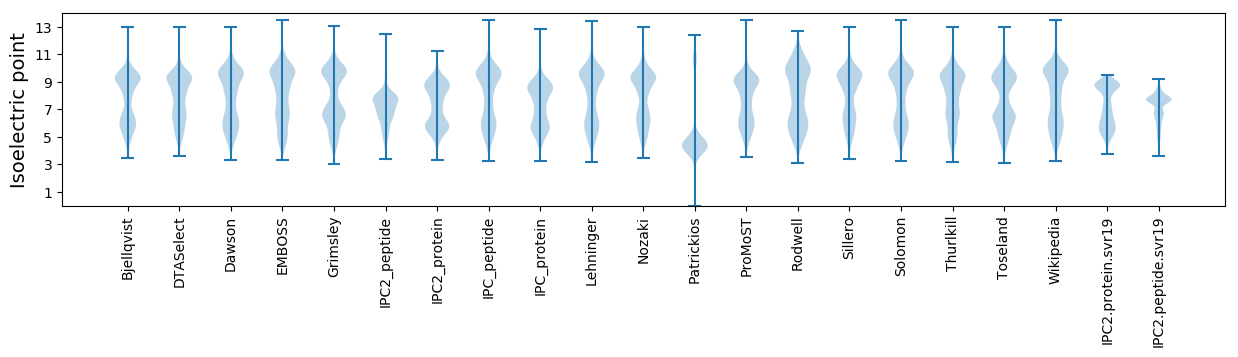
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X4XXY5|A0A1X4XXY5_9DELT Z1 domain-containing protein OS=Desulfurella amilsii OX=1562698 GN=DESAMIL20_1953 PE=4 SV=1
MM1 pKa = 7.38KK2 pKa = 9.6RR3 pKa = 11.84TFQPHH8 pKa = 4.18NTPRR12 pKa = 11.84KK13 pKa = 7.34RR14 pKa = 11.84THH16 pKa = 6.22GFRR19 pKa = 11.84TRR21 pKa = 11.84MKK23 pKa = 8.83TANGRR28 pKa = 11.84QVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.98GRR39 pKa = 11.84KK40 pKa = 8.8RR41 pKa = 11.84LAII44 pKa = 4.0
MM1 pKa = 7.38KK2 pKa = 9.6RR3 pKa = 11.84TFQPHH8 pKa = 4.18NTPRR12 pKa = 11.84KK13 pKa = 7.34RR14 pKa = 11.84THH16 pKa = 6.22GFRR19 pKa = 11.84TRR21 pKa = 11.84MKK23 pKa = 8.83TANGRR28 pKa = 11.84QVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.98GRR39 pKa = 11.84KK40 pKa = 8.8RR41 pKa = 11.84LAII44 pKa = 4.0
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
603201 |
37 |
1518 |
298.8 |
33.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.536 ± 0.045 | 1.071 ± 0.023 |
5.13 ± 0.04 | 6.04 ± 0.064 |
5.371 ± 0.057 | 5.905 ± 0.05 |
1.519 ± 0.02 | 9.505 ± 0.055 |
9.021 ± 0.06 | 9.813 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.116 ± 0.02 | 6.038 ± 0.054 |
3.295 ± 0.031 | 3.202 ± 0.039 |
3.015 ± 0.032 | 6.534 ± 0.047 |
4.769 ± 0.043 | 6.248 ± 0.036 |
0.698 ± 0.016 | 4.173 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
