
Beihai noda-like virus 11
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.19
Get precalculated fractions of proteins
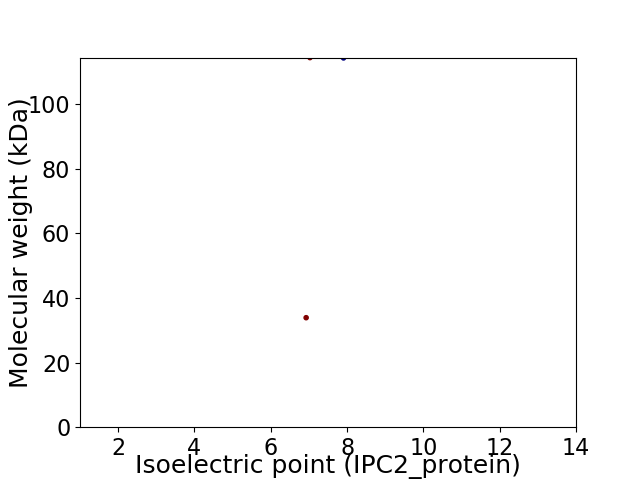
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFH1|A0A1L3KFH1_9VIRU Capsid protein OS=Beihai noda-like virus 11 OX=1922464 PE=4 SV=1
MM1 pKa = 7.55AKK3 pKa = 10.23KK4 pKa = 8.51PTKK7 pKa = 10.18KK8 pKa = 9.82NARR11 pKa = 11.84NRR13 pKa = 11.84GSNRR17 pKa = 11.84PRR19 pKa = 11.84GAPVAQATNIHH30 pKa = 6.09VGSRR34 pKa = 11.84DD35 pKa = 3.35PPIVVRR41 pKa = 11.84GTGRR45 pKa = 11.84FAHH48 pKa = 5.94STHH51 pKa = 6.74AANSLSDD58 pKa = 3.5GDD60 pKa = 4.15VLFDD64 pKa = 4.98SIINPSSFPRR74 pKa = 11.84MSRR77 pKa = 11.84IASAYY82 pKa = 7.76QRR84 pKa = 11.84YY85 pKa = 8.63RR86 pKa = 11.84FTRR89 pKa = 11.84LAFTIQPMCPATTGAGYY106 pKa = 10.56VAGFLKK112 pKa = 10.79DD113 pKa = 3.5PTDD116 pKa = 3.56EE117 pKa = 4.44DD118 pKa = 4.19TSFDD122 pKa = 4.8AIQGSQGAVIGKK134 pKa = 7.18WWEE137 pKa = 3.79HH138 pKa = 4.81KK139 pKa = 10.11RR140 pKa = 11.84IEE142 pKa = 4.31VRR144 pKa = 11.84PPTDD148 pKa = 4.24LLWTSLGEE156 pKa = 4.08NPRR159 pKa = 11.84LFSPGKK165 pKa = 9.95FVMTAVGTNTDD176 pKa = 3.3AVNVSVLCEE185 pKa = 3.98WEE187 pKa = 4.21AVLSVPSLEE196 pKa = 5.08DD197 pKa = 3.49YY198 pKa = 10.88SEE200 pKa = 4.86KK201 pKa = 10.61PVTEE205 pKa = 4.09YY206 pKa = 11.08FSTKK210 pKa = 10.61DD211 pKa = 3.82LLNSSYY217 pKa = 10.31VHH219 pKa = 6.43NSNSILQTNVFDD231 pKa = 4.98GDD233 pKa = 4.08VIPTGTILRR242 pKa = 11.84IPFPIDD248 pKa = 2.71VDD250 pKa = 3.91YY251 pKa = 11.64SLGAGDD257 pKa = 3.73VASARR262 pKa = 11.84YY263 pKa = 9.45CYY265 pKa = 10.28FLAGSHH271 pKa = 5.82NLTWGLYY278 pKa = 9.67SGGSFSGGSNYY289 pKa = 10.09YY290 pKa = 10.63SDD292 pKa = 4.45GQPTQCVLPKK302 pKa = 9.0GTRR305 pKa = 11.84FEE307 pKa = 4.64VIPNN311 pKa = 3.7
MM1 pKa = 7.55AKK3 pKa = 10.23KK4 pKa = 8.51PTKK7 pKa = 10.18KK8 pKa = 9.82NARR11 pKa = 11.84NRR13 pKa = 11.84GSNRR17 pKa = 11.84PRR19 pKa = 11.84GAPVAQATNIHH30 pKa = 6.09VGSRR34 pKa = 11.84DD35 pKa = 3.35PPIVVRR41 pKa = 11.84GTGRR45 pKa = 11.84FAHH48 pKa = 5.94STHH51 pKa = 6.74AANSLSDD58 pKa = 3.5GDD60 pKa = 4.15VLFDD64 pKa = 4.98SIINPSSFPRR74 pKa = 11.84MSRR77 pKa = 11.84IASAYY82 pKa = 7.76QRR84 pKa = 11.84YY85 pKa = 8.63RR86 pKa = 11.84FTRR89 pKa = 11.84LAFTIQPMCPATTGAGYY106 pKa = 10.56VAGFLKK112 pKa = 10.79DD113 pKa = 3.5PTDD116 pKa = 3.56EE117 pKa = 4.44DD118 pKa = 4.19TSFDD122 pKa = 4.8AIQGSQGAVIGKK134 pKa = 7.18WWEE137 pKa = 3.79HH138 pKa = 4.81KK139 pKa = 10.11RR140 pKa = 11.84IEE142 pKa = 4.31VRR144 pKa = 11.84PPTDD148 pKa = 4.24LLWTSLGEE156 pKa = 4.08NPRR159 pKa = 11.84LFSPGKK165 pKa = 9.95FVMTAVGTNTDD176 pKa = 3.3AVNVSVLCEE185 pKa = 3.98WEE187 pKa = 4.21AVLSVPSLEE196 pKa = 5.08DD197 pKa = 3.49YY198 pKa = 10.88SEE200 pKa = 4.86KK201 pKa = 10.61PVTEE205 pKa = 4.09YY206 pKa = 11.08FSTKK210 pKa = 10.61DD211 pKa = 3.82LLNSSYY217 pKa = 10.31VHH219 pKa = 6.43NSNSILQTNVFDD231 pKa = 4.98GDD233 pKa = 4.08VIPTGTILRR242 pKa = 11.84IPFPIDD248 pKa = 2.71VDD250 pKa = 3.91YY251 pKa = 11.64SLGAGDD257 pKa = 3.73VASARR262 pKa = 11.84YY263 pKa = 9.45CYY265 pKa = 10.28FLAGSHH271 pKa = 5.82NLTWGLYY278 pKa = 9.67SGGSFSGGSNYY289 pKa = 10.09YY290 pKa = 10.63SDD292 pKa = 4.45GQPTQCVLPKK302 pKa = 9.0GTRR305 pKa = 11.84FEE307 pKa = 4.64VIPNN311 pKa = 3.7
Molecular weight: 33.89 kDa
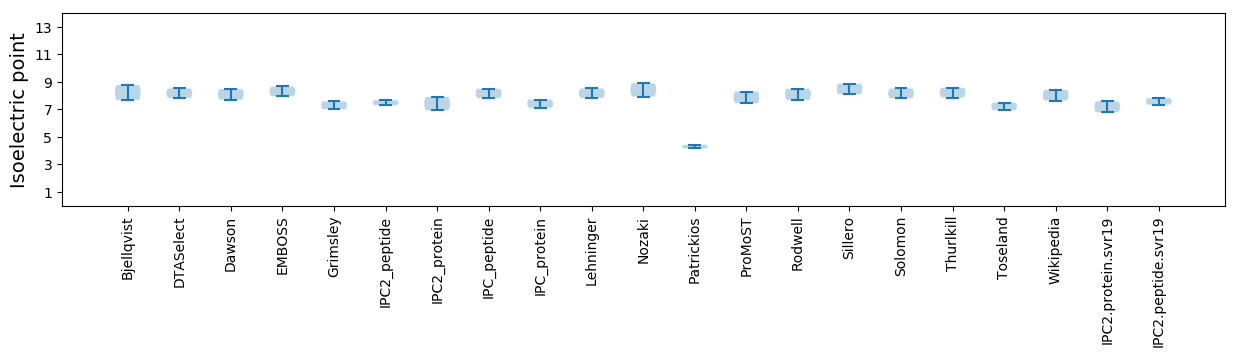
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFH1|A0A1L3KFH1_9VIRU Capsid protein OS=Beihai noda-like virus 11 OX=1922464 PE=4 SV=1
MM1 pKa = 7.45SLFHH5 pKa = 7.01FIASTFRR12 pKa = 11.84QGVFRR17 pKa = 11.84MSQSCLLWLITAIASVIGEE36 pKa = 4.31TLNLPVLIFISLAVFSLSIVAVVKK60 pKa = 9.14MARR63 pKa = 11.84SDD65 pKa = 3.09WFANRR70 pKa = 11.84LLDD73 pKa = 5.27LRR75 pKa = 11.84HH76 pKa = 5.53QLIGSSVIKK85 pKa = 10.53IPTASHH91 pKa = 6.48LRR93 pKa = 11.84STVQKK98 pKa = 9.68KK99 pKa = 10.75LEE101 pKa = 3.9MTKK104 pKa = 9.92CGVRR108 pKa = 11.84RR109 pKa = 11.84GHH111 pKa = 5.81SHH113 pKa = 4.72QQAATEE119 pKa = 4.26RR120 pKa = 11.84NSATEE125 pKa = 3.83TMLDD129 pKa = 3.8FVTSNGYY136 pKa = 8.87IPYY139 pKa = 9.8VISPSPRR146 pKa = 11.84EE147 pKa = 3.75AGIDD151 pKa = 3.43GRR153 pKa = 11.84RR154 pKa = 11.84RR155 pKa = 11.84FYY157 pKa = 11.46SLADD161 pKa = 3.38LRR163 pKa = 11.84QNYY166 pKa = 10.16LNDD169 pKa = 4.15PLTDD173 pKa = 3.03KK174 pKa = 10.92HH175 pKa = 8.12IIVMTDD181 pKa = 1.86VDD183 pKa = 4.78YY184 pKa = 11.13YY185 pKa = 11.75VNMHH189 pKa = 6.02EE190 pKa = 5.12LISLGRR196 pKa = 11.84PILCYY201 pKa = 9.63TFQPSTVSGPVKK213 pKa = 10.54DD214 pKa = 4.49GYY216 pKa = 9.65FTIKK220 pKa = 10.6DD221 pKa = 3.92DD222 pKa = 3.74VVHH225 pKa = 5.47YY226 pKa = 10.02HH227 pKa = 5.72VNGGKK232 pKa = 9.77DD233 pKa = 3.32VQHH236 pKa = 7.07RR237 pKa = 11.84IWNYY241 pKa = 9.09NQDD244 pKa = 3.64TIYY247 pKa = 11.22VVDD250 pKa = 4.29PEE252 pKa = 4.28NGFWATLRR260 pKa = 11.84QIIYY264 pKa = 10.55DD265 pKa = 3.87LVGLSWLSSICHH277 pKa = 5.62KK278 pKa = 10.63RR279 pKa = 11.84FGIGPFGRR287 pKa = 11.84KK288 pKa = 7.21VTICTVDD295 pKa = 3.17QFKK298 pKa = 11.14LSDD301 pKa = 3.31HH302 pKa = 6.63RR303 pKa = 11.84NIVSIVPFAKK313 pKa = 10.13CRR315 pKa = 11.84EE316 pKa = 3.95NLLPFEE322 pKa = 5.24EE323 pKa = 4.8YY324 pKa = 10.47GQQLRR329 pKa = 11.84RR330 pKa = 11.84MQYY333 pKa = 9.13GAANPTEE340 pKa = 4.12QFNTLIYY347 pKa = 10.71LGDD350 pKa = 4.04GDD352 pKa = 4.79PLVSLGVAGQVASVQLPLRR371 pKa = 11.84DD372 pKa = 4.1LEE374 pKa = 5.49AMMLMHH380 pKa = 7.18NDD382 pKa = 3.05SKK384 pKa = 10.14TANLSDD390 pKa = 3.43TVRR393 pKa = 11.84RR394 pKa = 11.84ARR396 pKa = 11.84ATEE399 pKa = 3.75KK400 pKa = 10.94SLTEE404 pKa = 3.59QQAAIVHH411 pKa = 6.4HH412 pKa = 6.64YY413 pKa = 9.11LLSMLPLKK421 pKa = 10.32PDD423 pKa = 3.43VVHH426 pKa = 7.27KK427 pKa = 10.24PGQLARR433 pKa = 11.84HH434 pKa = 5.07YY435 pKa = 10.45QATDD439 pKa = 2.74VKK441 pKa = 10.41YY442 pKa = 11.02DD443 pKa = 3.73HH444 pKa = 7.72DD445 pKa = 3.84CTEE448 pKa = 3.69QGRR451 pKa = 11.84EE452 pKa = 3.9YY453 pKa = 11.13ARR455 pKa = 11.84TYY457 pKa = 11.39APGPLSQTAVFPNEE471 pKa = 3.28SRR473 pKa = 11.84ANEE476 pKa = 3.93RR477 pKa = 11.84ATIDD481 pKa = 3.42GRR483 pKa = 11.84VVGPQTKK490 pKa = 10.24AKK492 pKa = 10.0ARR494 pKa = 11.84EE495 pKa = 4.23NITQRR500 pKa = 11.84LRR502 pKa = 11.84KK503 pKa = 8.28YY504 pKa = 10.69AKK506 pKa = 10.41DD507 pKa = 3.46FVKK510 pKa = 10.62QLVPNPGAGFRR521 pKa = 11.84YY522 pKa = 9.35SVAHH526 pKa = 6.6VEE528 pKa = 4.23EE529 pKa = 4.59QQQKK533 pKa = 8.97PLQRR537 pKa = 11.84ARR539 pKa = 11.84NDD541 pKa = 3.38ANRR544 pKa = 11.84MHH546 pKa = 6.8HH547 pKa = 7.34AISMITKK554 pKa = 9.73SFQKK558 pKa = 10.4KK559 pKa = 7.19EE560 pKa = 3.8AYY562 pKa = 8.38GAPNHH567 pKa = 6.41PRR569 pKa = 11.84NISTVPHH576 pKa = 5.77GQNARR581 pKa = 11.84LSGYY585 pKa = 8.89TYY587 pKa = 10.94AFKK590 pKa = 10.91DD591 pKa = 3.46AVLKK595 pKa = 8.91KK596 pKa = 8.52TEE598 pKa = 3.81WYY600 pKa = 8.39MPCKK604 pKa = 9.91TPAQIAEE611 pKa = 4.27AVVNLAAKK619 pKa = 8.93STEE622 pKa = 4.03LVEE625 pKa = 4.1TDD627 pKa = 3.26YY628 pKa = 11.77SRR630 pKa = 11.84FDD632 pKa = 3.3GTFLRR637 pKa = 11.84FMRR640 pKa = 11.84EE641 pKa = 3.82HH642 pKa = 8.3VEE644 pKa = 4.0FACYY648 pKa = 9.55KK649 pKa = 9.01RR650 pKa = 11.84WVPEE654 pKa = 3.63EE655 pKa = 4.1LLPEE659 pKa = 4.48LSEE662 pKa = 4.13LLNNEE667 pKa = 3.94VDD669 pKa = 3.64SKK671 pKa = 11.66ARR673 pKa = 11.84TRR675 pKa = 11.84LGLTYY680 pKa = 10.78APGCSRR686 pKa = 11.84LSGSPLTTDD695 pKa = 3.96GNSICNAFVSYY706 pKa = 10.76AAGRR710 pKa = 11.84YY711 pKa = 9.04SGASHH716 pKa = 6.77EE717 pKa = 4.65EE718 pKa = 3.63AWDD721 pKa = 4.57DD722 pKa = 3.36IGIVYY727 pKa = 10.4GDD729 pKa = 3.96DD730 pKa = 3.65GLRR733 pKa = 11.84SGSVPDD739 pKa = 4.13EE740 pKa = 4.46LLSSTASDD748 pKa = 3.8LGFEE752 pKa = 4.81LKK754 pKa = 9.92ICNRR758 pKa = 11.84ASRR761 pKa = 11.84GQSVSFLSRR770 pKa = 11.84IFCDD774 pKa = 3.36PWSSPASIQDD784 pKa = 3.61PARR787 pKa = 11.84TLLKK791 pKa = 10.33IHH793 pKa = 5.84TTCDD797 pKa = 3.21GSNKK801 pKa = 9.29TIQEE805 pKa = 3.97IGWAKK810 pKa = 7.63TQAYY814 pKa = 8.91LATDD818 pKa = 3.97RR819 pKa = 11.84LTPFISDD826 pKa = 2.71WCLAYY831 pKa = 10.31QRR833 pKa = 11.84NCPEE837 pKa = 3.62QLVNYY842 pKa = 9.98DD843 pKa = 4.91DD844 pKa = 4.09YY845 pKa = 11.8TDD847 pKa = 3.21IPFWVRR853 pKa = 11.84DD854 pKa = 3.56EE855 pKa = 5.67DD856 pKa = 4.06SLNNSWPQSDD866 pKa = 3.63NEE868 pKa = 3.87IWVGIVANRR877 pKa = 11.84LGVTASEE884 pKa = 4.18LDD886 pKa = 3.34AHH888 pKa = 6.39IKK890 pKa = 9.82KK891 pKa = 10.15LRR893 pKa = 11.84AFTGRR898 pKa = 11.84VEE900 pKa = 4.51DD901 pKa = 4.34LPVLTSHH908 pKa = 7.76IDD910 pKa = 3.32QTPKK914 pKa = 11.02LEE916 pKa = 3.91VAMDD920 pKa = 4.03GEE922 pKa = 4.56IHH924 pKa = 7.07AGPSSVNKK932 pKa = 9.02KK933 pKa = 8.19TEE935 pKa = 3.87NGPNHH940 pKa = 6.53HH941 pKa = 7.19GDD943 pKa = 3.73RR944 pKa = 11.84EE945 pKa = 4.19DD946 pKa = 3.55SGTAEE951 pKa = 4.18TAVPTSRR958 pKa = 11.84GSPAKK963 pKa = 9.79PGRR966 pKa = 11.84AVPGRR971 pKa = 11.84GKK973 pKa = 10.39RR974 pKa = 11.84DD975 pKa = 3.38ANVCAEE981 pKa = 4.27RR982 pKa = 11.84EE983 pKa = 4.25RR984 pKa = 11.84KK985 pKa = 9.58GGRR988 pKa = 11.84NQASAIAHH996 pKa = 5.93SNSASADD1003 pKa = 3.4KK1004 pKa = 10.19PHH1006 pKa = 7.06RR1007 pKa = 11.84RR1008 pKa = 11.84RR1009 pKa = 11.84PTPQVSGPRR1018 pKa = 11.84KK1019 pKa = 9.2PKK1021 pKa = 9.94VV1022 pKa = 3.06
MM1 pKa = 7.45SLFHH5 pKa = 7.01FIASTFRR12 pKa = 11.84QGVFRR17 pKa = 11.84MSQSCLLWLITAIASVIGEE36 pKa = 4.31TLNLPVLIFISLAVFSLSIVAVVKK60 pKa = 9.14MARR63 pKa = 11.84SDD65 pKa = 3.09WFANRR70 pKa = 11.84LLDD73 pKa = 5.27LRR75 pKa = 11.84HH76 pKa = 5.53QLIGSSVIKK85 pKa = 10.53IPTASHH91 pKa = 6.48LRR93 pKa = 11.84STVQKK98 pKa = 9.68KK99 pKa = 10.75LEE101 pKa = 3.9MTKK104 pKa = 9.92CGVRR108 pKa = 11.84RR109 pKa = 11.84GHH111 pKa = 5.81SHH113 pKa = 4.72QQAATEE119 pKa = 4.26RR120 pKa = 11.84NSATEE125 pKa = 3.83TMLDD129 pKa = 3.8FVTSNGYY136 pKa = 8.87IPYY139 pKa = 9.8VISPSPRR146 pKa = 11.84EE147 pKa = 3.75AGIDD151 pKa = 3.43GRR153 pKa = 11.84RR154 pKa = 11.84RR155 pKa = 11.84FYY157 pKa = 11.46SLADD161 pKa = 3.38LRR163 pKa = 11.84QNYY166 pKa = 10.16LNDD169 pKa = 4.15PLTDD173 pKa = 3.03KK174 pKa = 10.92HH175 pKa = 8.12IIVMTDD181 pKa = 1.86VDD183 pKa = 4.78YY184 pKa = 11.13YY185 pKa = 11.75VNMHH189 pKa = 6.02EE190 pKa = 5.12LISLGRR196 pKa = 11.84PILCYY201 pKa = 9.63TFQPSTVSGPVKK213 pKa = 10.54DD214 pKa = 4.49GYY216 pKa = 9.65FTIKK220 pKa = 10.6DD221 pKa = 3.92DD222 pKa = 3.74VVHH225 pKa = 5.47YY226 pKa = 10.02HH227 pKa = 5.72VNGGKK232 pKa = 9.77DD233 pKa = 3.32VQHH236 pKa = 7.07RR237 pKa = 11.84IWNYY241 pKa = 9.09NQDD244 pKa = 3.64TIYY247 pKa = 11.22VVDD250 pKa = 4.29PEE252 pKa = 4.28NGFWATLRR260 pKa = 11.84QIIYY264 pKa = 10.55DD265 pKa = 3.87LVGLSWLSSICHH277 pKa = 5.62KK278 pKa = 10.63RR279 pKa = 11.84FGIGPFGRR287 pKa = 11.84KK288 pKa = 7.21VTICTVDD295 pKa = 3.17QFKK298 pKa = 11.14LSDD301 pKa = 3.31HH302 pKa = 6.63RR303 pKa = 11.84NIVSIVPFAKK313 pKa = 10.13CRR315 pKa = 11.84EE316 pKa = 3.95NLLPFEE322 pKa = 5.24EE323 pKa = 4.8YY324 pKa = 10.47GQQLRR329 pKa = 11.84RR330 pKa = 11.84MQYY333 pKa = 9.13GAANPTEE340 pKa = 4.12QFNTLIYY347 pKa = 10.71LGDD350 pKa = 4.04GDD352 pKa = 4.79PLVSLGVAGQVASVQLPLRR371 pKa = 11.84DD372 pKa = 4.1LEE374 pKa = 5.49AMMLMHH380 pKa = 7.18NDD382 pKa = 3.05SKK384 pKa = 10.14TANLSDD390 pKa = 3.43TVRR393 pKa = 11.84RR394 pKa = 11.84ARR396 pKa = 11.84ATEE399 pKa = 3.75KK400 pKa = 10.94SLTEE404 pKa = 3.59QQAAIVHH411 pKa = 6.4HH412 pKa = 6.64YY413 pKa = 9.11LLSMLPLKK421 pKa = 10.32PDD423 pKa = 3.43VVHH426 pKa = 7.27KK427 pKa = 10.24PGQLARR433 pKa = 11.84HH434 pKa = 5.07YY435 pKa = 10.45QATDD439 pKa = 2.74VKK441 pKa = 10.41YY442 pKa = 11.02DD443 pKa = 3.73HH444 pKa = 7.72DD445 pKa = 3.84CTEE448 pKa = 3.69QGRR451 pKa = 11.84EE452 pKa = 3.9YY453 pKa = 11.13ARR455 pKa = 11.84TYY457 pKa = 11.39APGPLSQTAVFPNEE471 pKa = 3.28SRR473 pKa = 11.84ANEE476 pKa = 3.93RR477 pKa = 11.84ATIDD481 pKa = 3.42GRR483 pKa = 11.84VVGPQTKK490 pKa = 10.24AKK492 pKa = 10.0ARR494 pKa = 11.84EE495 pKa = 4.23NITQRR500 pKa = 11.84LRR502 pKa = 11.84KK503 pKa = 8.28YY504 pKa = 10.69AKK506 pKa = 10.41DD507 pKa = 3.46FVKK510 pKa = 10.62QLVPNPGAGFRR521 pKa = 11.84YY522 pKa = 9.35SVAHH526 pKa = 6.6VEE528 pKa = 4.23EE529 pKa = 4.59QQQKK533 pKa = 8.97PLQRR537 pKa = 11.84ARR539 pKa = 11.84NDD541 pKa = 3.38ANRR544 pKa = 11.84MHH546 pKa = 6.8HH547 pKa = 7.34AISMITKK554 pKa = 9.73SFQKK558 pKa = 10.4KK559 pKa = 7.19EE560 pKa = 3.8AYY562 pKa = 8.38GAPNHH567 pKa = 6.41PRR569 pKa = 11.84NISTVPHH576 pKa = 5.77GQNARR581 pKa = 11.84LSGYY585 pKa = 8.89TYY587 pKa = 10.94AFKK590 pKa = 10.91DD591 pKa = 3.46AVLKK595 pKa = 8.91KK596 pKa = 8.52TEE598 pKa = 3.81WYY600 pKa = 8.39MPCKK604 pKa = 9.91TPAQIAEE611 pKa = 4.27AVVNLAAKK619 pKa = 8.93STEE622 pKa = 4.03LVEE625 pKa = 4.1TDD627 pKa = 3.26YY628 pKa = 11.77SRR630 pKa = 11.84FDD632 pKa = 3.3GTFLRR637 pKa = 11.84FMRR640 pKa = 11.84EE641 pKa = 3.82HH642 pKa = 8.3VEE644 pKa = 4.0FACYY648 pKa = 9.55KK649 pKa = 9.01RR650 pKa = 11.84WVPEE654 pKa = 3.63EE655 pKa = 4.1LLPEE659 pKa = 4.48LSEE662 pKa = 4.13LLNNEE667 pKa = 3.94VDD669 pKa = 3.64SKK671 pKa = 11.66ARR673 pKa = 11.84TRR675 pKa = 11.84LGLTYY680 pKa = 10.78APGCSRR686 pKa = 11.84LSGSPLTTDD695 pKa = 3.96GNSICNAFVSYY706 pKa = 10.76AAGRR710 pKa = 11.84YY711 pKa = 9.04SGASHH716 pKa = 6.77EE717 pKa = 4.65EE718 pKa = 3.63AWDD721 pKa = 4.57DD722 pKa = 3.36IGIVYY727 pKa = 10.4GDD729 pKa = 3.96DD730 pKa = 3.65GLRR733 pKa = 11.84SGSVPDD739 pKa = 4.13EE740 pKa = 4.46LLSSTASDD748 pKa = 3.8LGFEE752 pKa = 4.81LKK754 pKa = 9.92ICNRR758 pKa = 11.84ASRR761 pKa = 11.84GQSVSFLSRR770 pKa = 11.84IFCDD774 pKa = 3.36PWSSPASIQDD784 pKa = 3.61PARR787 pKa = 11.84TLLKK791 pKa = 10.33IHH793 pKa = 5.84TTCDD797 pKa = 3.21GSNKK801 pKa = 9.29TIQEE805 pKa = 3.97IGWAKK810 pKa = 7.63TQAYY814 pKa = 8.91LATDD818 pKa = 3.97RR819 pKa = 11.84LTPFISDD826 pKa = 2.71WCLAYY831 pKa = 10.31QRR833 pKa = 11.84NCPEE837 pKa = 3.62QLVNYY842 pKa = 9.98DD843 pKa = 4.91DD844 pKa = 4.09YY845 pKa = 11.8TDD847 pKa = 3.21IPFWVRR853 pKa = 11.84DD854 pKa = 3.56EE855 pKa = 5.67DD856 pKa = 4.06SLNNSWPQSDD866 pKa = 3.63NEE868 pKa = 3.87IWVGIVANRR877 pKa = 11.84LGVTASEE884 pKa = 4.18LDD886 pKa = 3.34AHH888 pKa = 6.39IKK890 pKa = 9.82KK891 pKa = 10.15LRR893 pKa = 11.84AFTGRR898 pKa = 11.84VEE900 pKa = 4.51DD901 pKa = 4.34LPVLTSHH908 pKa = 7.76IDD910 pKa = 3.32QTPKK914 pKa = 11.02LEE916 pKa = 3.91VAMDD920 pKa = 4.03GEE922 pKa = 4.56IHH924 pKa = 7.07AGPSSVNKK932 pKa = 9.02KK933 pKa = 8.19TEE935 pKa = 3.87NGPNHH940 pKa = 6.53HH941 pKa = 7.19GDD943 pKa = 3.73RR944 pKa = 11.84EE945 pKa = 4.19DD946 pKa = 3.55SGTAEE951 pKa = 4.18TAVPTSRR958 pKa = 11.84GSPAKK963 pKa = 9.79PGRR966 pKa = 11.84AVPGRR971 pKa = 11.84GKK973 pKa = 10.39RR974 pKa = 11.84DD975 pKa = 3.38ANVCAEE981 pKa = 4.27RR982 pKa = 11.84EE983 pKa = 4.25RR984 pKa = 11.84KK985 pKa = 9.58GGRR988 pKa = 11.84NQASAIAHH996 pKa = 5.93SNSASADD1003 pKa = 3.4KK1004 pKa = 10.19PHH1006 pKa = 7.06RR1007 pKa = 11.84RR1008 pKa = 11.84RR1009 pKa = 11.84PTPQVSGPRR1018 pKa = 11.84KK1019 pKa = 9.2PKK1021 pKa = 9.94VV1022 pKa = 3.06
Molecular weight: 114.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1333 |
311 |
1022 |
666.5 |
74.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.952 ± 0.268 | 1.575 ± 0.139 |
5.851 ± 0.031 | 4.501 ± 0.62 |
3.676 ± 0.553 | 6.827 ± 0.894 |
2.926 ± 0.48 | 5.026 ± 0.098 |
4.501 ± 0.465 | 7.727 ± 0.625 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.575 ± 0.139 | 4.576 ± 0.274 |
5.851 ± 0.589 | 3.901 ± 0.64 |
6.827 ± 0.501 | 8.327 ± 0.791 |
6.377 ± 0.491 | 6.977 ± 0.202 |
1.425 ± 0.088 | 3.601 ± 0.124 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
