
Ajellomyces capsulatus (strain H88) (Darling s disease fungus) (Histoplasma capsulatum)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Onygenales; Ajellomycetaceae; Histoplasma; Histoplasma capsulatum
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
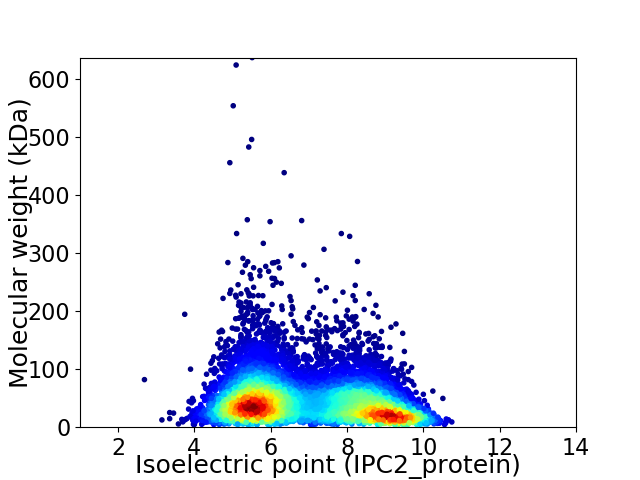
Virtual 2D-PAGE plot for 9445 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
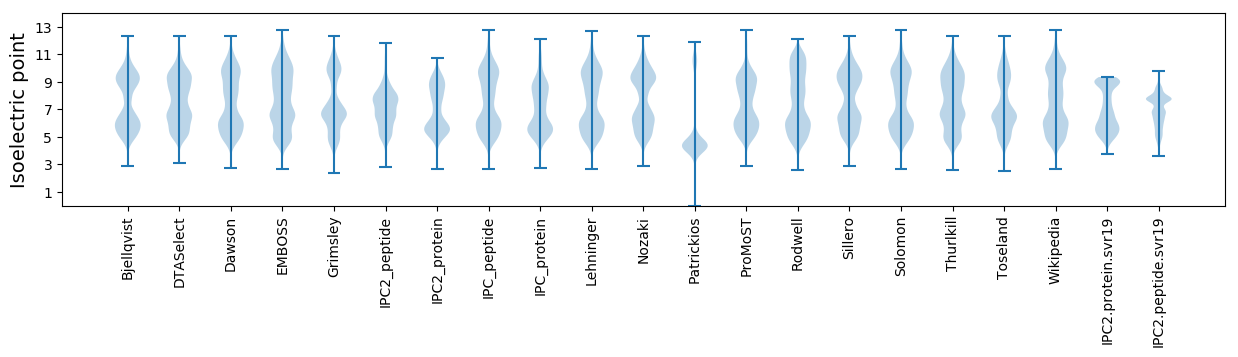
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|F0UHR3|F0UHR3_AJEC8 Patatin-like phospholipase domain-containing protein OS=Ajellomyces capsulatus (strain H88) OX=544711 GN=HCEG_05487 PE=3 SV=1
MM1 pKa = 7.67FSLTDD6 pKa = 4.04LSAADD11 pKa = 4.05LDD13 pKa = 4.39YY14 pKa = 11.0FSSHH18 pKa = 7.54IILTVHH24 pKa = 6.36NADD27 pKa = 4.23AFAINNNDD35 pKa = 3.35ADD37 pKa = 3.54QSYY40 pKa = 11.06EE41 pKa = 4.0KK42 pKa = 10.66FII44 pKa = 5.05
MM1 pKa = 7.67FSLTDD6 pKa = 4.04LSAADD11 pKa = 4.05LDD13 pKa = 4.39YY14 pKa = 11.0FSSHH18 pKa = 7.54IILTVHH24 pKa = 6.36NADD27 pKa = 4.23AFAINNNDD35 pKa = 3.35ADD37 pKa = 3.54QSYY40 pKa = 11.06EE41 pKa = 4.0KK42 pKa = 10.66FII44 pKa = 5.05
Molecular weight: 4.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F0UV39|F0UV39_AJEC8 Uncharacterized protein OS=Ajellomyces capsulatus (strain H88) OX=544711 GN=HCEG_08981 PE=4 SV=1
MM1 pKa = 7.5NVRR4 pKa = 11.84CLQVGIHH11 pKa = 6.15GFININISARR21 pKa = 11.84ANRR24 pKa = 11.84RR25 pKa = 11.84SSPSKK30 pKa = 9.81LSVYY34 pKa = 7.01VQCVQAYY41 pKa = 7.43PPPTLLSSVAYY52 pKa = 8.82PRR54 pKa = 11.84TIIPTTSAKK63 pKa = 10.42GRR65 pKa = 11.84DD66 pKa = 3.59IVLKK70 pKa = 10.31RR71 pKa = 11.84APEE74 pKa = 4.29RR75 pKa = 11.84DD76 pKa = 3.72DD77 pKa = 5.63KK78 pKa = 11.35ILEE81 pKa = 4.24TDD83 pKa = 3.04HH84 pKa = 7.41DD85 pKa = 4.18IPRR88 pKa = 11.84PSQGLLPSPSGAWHH102 pKa = 7.25PSQICQSSWAHH113 pKa = 6.7SNFQGCSIHH122 pKa = 6.38SQAARR127 pKa = 11.84DD128 pKa = 3.72VARR131 pKa = 11.84LKK133 pKa = 10.35IHH135 pKa = 6.01VGKK138 pKa = 8.49TRR140 pKa = 11.84WNGQEE145 pKa = 3.23IDD147 pKa = 3.36PRR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84GAQHH155 pKa = 6.41SFSLVHH161 pKa = 6.14NRR163 pKa = 11.84HH164 pKa = 5.62GGCWDD169 pKa = 3.32RR170 pKa = 11.84TTCRR174 pKa = 11.84MPQDD178 pKa = 3.53AKK180 pKa = 10.8SISSARR186 pKa = 11.84SHH188 pKa = 7.48PIPATEE194 pKa = 3.98RR195 pKa = 11.84ADD197 pKa = 3.53VPVGIVVYY205 pKa = 7.21EE206 pKa = 3.97TRR208 pKa = 11.84PQHH211 pKa = 6.23WDD213 pKa = 3.01PAAEE217 pKa = 4.01RR218 pKa = 11.84GSRR221 pKa = 11.84GWEE224 pKa = 3.78TKK226 pKa = 10.46VIEE229 pKa = 4.66GSQPQRR235 pKa = 11.84KK236 pKa = 8.16FFNFSVPTTHH246 pKa = 7.5RR247 pKa = 11.84VGIPLHH253 pKa = 6.27ARR255 pKa = 11.84AILAIIHH262 pKa = 5.71RR263 pKa = 11.84TFFLQNNPLLLASPP277 pKa = 4.76
MM1 pKa = 7.5NVRR4 pKa = 11.84CLQVGIHH11 pKa = 6.15GFININISARR21 pKa = 11.84ANRR24 pKa = 11.84RR25 pKa = 11.84SSPSKK30 pKa = 9.81LSVYY34 pKa = 7.01VQCVQAYY41 pKa = 7.43PPPTLLSSVAYY52 pKa = 8.82PRR54 pKa = 11.84TIIPTTSAKK63 pKa = 10.42GRR65 pKa = 11.84DD66 pKa = 3.59IVLKK70 pKa = 10.31RR71 pKa = 11.84APEE74 pKa = 4.29RR75 pKa = 11.84DD76 pKa = 3.72DD77 pKa = 5.63KK78 pKa = 11.35ILEE81 pKa = 4.24TDD83 pKa = 3.04HH84 pKa = 7.41DD85 pKa = 4.18IPRR88 pKa = 11.84PSQGLLPSPSGAWHH102 pKa = 7.25PSQICQSSWAHH113 pKa = 6.7SNFQGCSIHH122 pKa = 6.38SQAARR127 pKa = 11.84DD128 pKa = 3.72VARR131 pKa = 11.84LKK133 pKa = 10.35IHH135 pKa = 6.01VGKK138 pKa = 8.49TRR140 pKa = 11.84WNGQEE145 pKa = 3.23IDD147 pKa = 3.36PRR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84GAQHH155 pKa = 6.41SFSLVHH161 pKa = 6.14NRR163 pKa = 11.84HH164 pKa = 5.62GGCWDD169 pKa = 3.32RR170 pKa = 11.84TTCRR174 pKa = 11.84MPQDD178 pKa = 3.53AKK180 pKa = 10.8SISSARR186 pKa = 11.84SHH188 pKa = 7.48PIPATEE194 pKa = 3.98RR195 pKa = 11.84ADD197 pKa = 3.53VPVGIVVYY205 pKa = 7.21EE206 pKa = 3.97TRR208 pKa = 11.84PQHH211 pKa = 6.23WDD213 pKa = 3.01PAAEE217 pKa = 4.01RR218 pKa = 11.84GSRR221 pKa = 11.84GWEE224 pKa = 3.78TKK226 pKa = 10.46VIEE229 pKa = 4.66GSQPQRR235 pKa = 11.84KK236 pKa = 8.16FFNFSVPTTHH246 pKa = 7.5RR247 pKa = 11.84VGIPLHH253 pKa = 6.27ARR255 pKa = 11.84AILAIIHH262 pKa = 5.71RR263 pKa = 11.84TFFLQNNPLLLASPP277 pKa = 4.76
Molecular weight: 30.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4200093 |
39 |
5717 |
444.7 |
49.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.907 ± 0.018 | 1.277 ± 0.009 |
5.54 ± 0.017 | 6.118 ± 0.026 |
3.642 ± 0.014 | 6.725 ± 0.023 |
2.522 ± 0.011 | 5.102 ± 0.016 |
4.996 ± 0.02 | 8.875 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.091 ± 0.009 | 3.967 ± 0.013 |
6.289 ± 0.029 | 4.075 ± 0.018 |
6.452 ± 0.021 | 8.831 ± 0.03 |
5.857 ± 0.016 | 5.787 ± 0.018 |
1.326 ± 0.009 | 2.62 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
