
Halovirus HCTV-2
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified archaeal dsDNA viruses; Haloviruses
Average proteome isoelectric point is 5.23
Get precalculated fractions of proteins
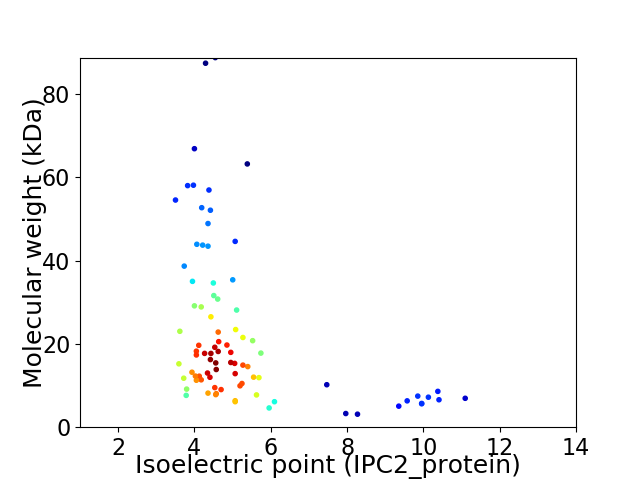
Virtual 2D-PAGE plot for 86 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R4TNM4|R4TNM4_9VIRU Uncharacterized protein OS=Halovirus HCTV-2 OX=1273747 GN=64 PE=4 SV=1
MM1 pKa = 7.49ALEE4 pKa = 4.88PIGDD8 pKa = 3.49IGVAALTEE16 pKa = 4.31QVGSEE21 pKa = 4.12TLTWATAAEE30 pKa = 3.87WDD32 pKa = 4.07ANVSEE37 pKa = 4.48TGVVHH42 pKa = 7.14EE43 pKa = 5.32DD44 pKa = 3.15FGDD47 pKa = 3.74RR48 pKa = 11.84DD49 pKa = 3.42ATRR52 pKa = 11.84IEE54 pKa = 4.44LGFPSTDD61 pKa = 3.23PVNSGLAYY69 pKa = 10.29YY70 pKa = 10.27YY71 pKa = 10.11PLDD74 pKa = 4.18EE75 pKa = 5.09ASGPATDD82 pKa = 3.91ATGNGPNGSLSGSTSLGNSGTLGTTCIDD110 pKa = 3.71FPGGGGDD117 pKa = 3.56TLDD120 pKa = 4.86FPDD123 pKa = 5.07DD124 pKa = 4.16PVWDD128 pKa = 3.5ITGDD132 pKa = 3.23ITVVHH137 pKa = 7.0AIYY140 pKa = 9.82WRR142 pKa = 11.84GGNDD146 pKa = 3.05SSSWQGLTSKK156 pKa = 10.58GGNSSPWAIAYY167 pKa = 10.02VDD169 pKa = 4.02GYY171 pKa = 10.54YY172 pKa = 10.49VQFQVDD178 pKa = 3.66GTRR181 pKa = 11.84YY182 pKa = 8.57DD183 pKa = 2.69TGYY186 pKa = 11.25DD187 pKa = 3.6LRR189 pKa = 11.84NTQNTWALHH198 pKa = 6.6AYY200 pKa = 9.63QLDD203 pKa = 3.93SGSIRR208 pKa = 11.84HH209 pKa = 5.21QVNGGNEE216 pKa = 4.0HH217 pKa = 7.09DD218 pKa = 3.96TTNSAEE224 pKa = 4.52AINNTTSDD232 pKa = 3.75EE233 pKa = 4.34FVLGARR239 pKa = 11.84KK240 pKa = 10.12SGGDD244 pKa = 3.51EE245 pKa = 3.95VNALVDD251 pKa = 3.62DD252 pKa = 4.73FFVFDD257 pKa = 3.93YY258 pKa = 10.97AISDD262 pKa = 3.54TRR264 pKa = 11.84LTDD267 pKa = 5.63LYY269 pKa = 10.82DD270 pKa = 3.27AWSTGTLTTATKK282 pKa = 9.28TVSSSGQPDD291 pKa = 4.03LQNLVYY297 pKa = 10.41SLNGEE302 pKa = 4.51SITLDD307 pKa = 3.54VIGSPGTASEE317 pKa = 4.29EE318 pKa = 4.55TVTQALDD325 pKa = 3.55GSSSYY330 pKa = 10.79TLTWADD336 pKa = 2.86SHH338 pKa = 6.05TDD340 pKa = 3.61FRR342 pKa = 11.84VRR344 pKa = 11.84AEE346 pKa = 4.42LTTSDD351 pKa = 3.79PTVSPTVSQIEE362 pKa = 4.39LVTT365 pKa = 3.71
MM1 pKa = 7.49ALEE4 pKa = 4.88PIGDD8 pKa = 3.49IGVAALTEE16 pKa = 4.31QVGSEE21 pKa = 4.12TLTWATAAEE30 pKa = 3.87WDD32 pKa = 4.07ANVSEE37 pKa = 4.48TGVVHH42 pKa = 7.14EE43 pKa = 5.32DD44 pKa = 3.15FGDD47 pKa = 3.74RR48 pKa = 11.84DD49 pKa = 3.42ATRR52 pKa = 11.84IEE54 pKa = 4.44LGFPSTDD61 pKa = 3.23PVNSGLAYY69 pKa = 10.29YY70 pKa = 10.27YY71 pKa = 10.11PLDD74 pKa = 4.18EE75 pKa = 5.09ASGPATDD82 pKa = 3.91ATGNGPNGSLSGSTSLGNSGTLGTTCIDD110 pKa = 3.71FPGGGGDD117 pKa = 3.56TLDD120 pKa = 4.86FPDD123 pKa = 5.07DD124 pKa = 4.16PVWDD128 pKa = 3.5ITGDD132 pKa = 3.23ITVVHH137 pKa = 7.0AIYY140 pKa = 9.82WRR142 pKa = 11.84GGNDD146 pKa = 3.05SSSWQGLTSKK156 pKa = 10.58GGNSSPWAIAYY167 pKa = 10.02VDD169 pKa = 4.02GYY171 pKa = 10.54YY172 pKa = 10.49VQFQVDD178 pKa = 3.66GTRR181 pKa = 11.84YY182 pKa = 8.57DD183 pKa = 2.69TGYY186 pKa = 11.25DD187 pKa = 3.6LRR189 pKa = 11.84NTQNTWALHH198 pKa = 6.6AYY200 pKa = 9.63QLDD203 pKa = 3.93SGSIRR208 pKa = 11.84HH209 pKa = 5.21QVNGGNEE216 pKa = 4.0HH217 pKa = 7.09DD218 pKa = 3.96TTNSAEE224 pKa = 4.52AINNTTSDD232 pKa = 3.75EE233 pKa = 4.34FVLGARR239 pKa = 11.84KK240 pKa = 10.12SGGDD244 pKa = 3.51EE245 pKa = 3.95VNALVDD251 pKa = 3.62DD252 pKa = 4.73FFVFDD257 pKa = 3.93YY258 pKa = 10.97AISDD262 pKa = 3.54TRR264 pKa = 11.84LTDD267 pKa = 5.63LYY269 pKa = 10.82DD270 pKa = 3.27AWSTGTLTTATKK282 pKa = 9.28TVSSSGQPDD291 pKa = 4.03LQNLVYY297 pKa = 10.41SLNGEE302 pKa = 4.51SITLDD307 pKa = 3.54VIGSPGTASEE317 pKa = 4.29EE318 pKa = 4.55TVTQALDD325 pKa = 3.55GSSSYY330 pKa = 10.79TLTWADD336 pKa = 2.86SHH338 pKa = 6.05TDD340 pKa = 3.61FRR342 pKa = 11.84VRR344 pKa = 11.84AEE346 pKa = 4.42LTTSDD351 pKa = 3.79PTVSPTVSQIEE362 pKa = 4.39LVTT365 pKa = 3.71
Molecular weight: 38.66 kDa
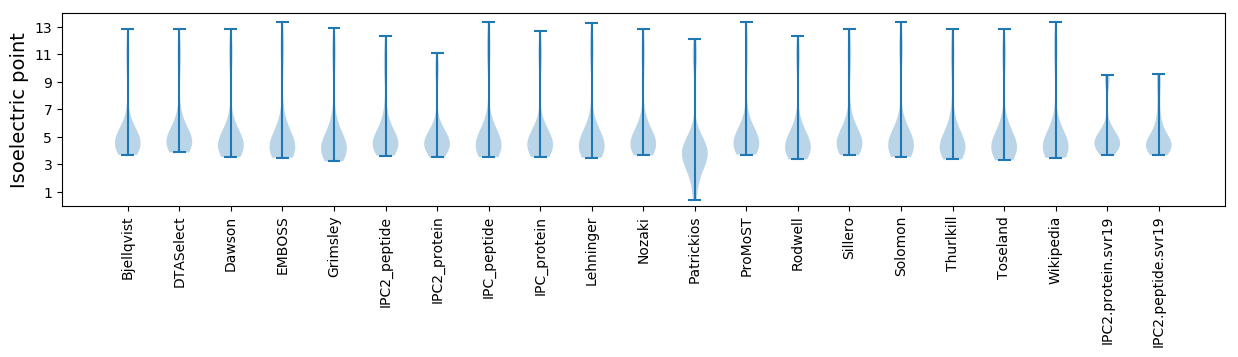
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R4TNQ4|R4TNQ4_9VIRU Uncharacterized protein OS=Halovirus HCTV-2 OX=1273747 GN=85 PE=4 SV=1
MM1 pKa = 7.64RR2 pKa = 11.84GARR5 pKa = 11.84PVRR8 pKa = 11.84TRR10 pKa = 11.84TRR12 pKa = 11.84SVTRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84SLVDD22 pKa = 3.33AGRR25 pKa = 11.84GRR27 pKa = 11.84LGRR30 pKa = 11.84ALRR33 pKa = 11.84PLAHH37 pKa = 7.37RR38 pKa = 11.84SPTTHH43 pKa = 7.92PLRR46 pKa = 11.84PPPSVRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84ALFSPFAA61 pKa = 5.27
MM1 pKa = 7.64RR2 pKa = 11.84GARR5 pKa = 11.84PVRR8 pKa = 11.84TRR10 pKa = 11.84TRR12 pKa = 11.84SVTRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84SLVDD22 pKa = 3.33AGRR25 pKa = 11.84GRR27 pKa = 11.84LGRR30 pKa = 11.84ALRR33 pKa = 11.84PLAHH37 pKa = 7.37RR38 pKa = 11.84SPTTHH43 pKa = 7.92PLRR46 pKa = 11.84PPPSVRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84ALFSPFAA61 pKa = 5.27
Molecular weight: 6.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
17670 |
31 |
786 |
205.5 |
22.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.655 ± 0.296 | 1.047 ± 0.161 |
8.149 ± 0.235 | 9.502 ± 0.436 |
2.315 ± 0.19 | 9.225 ± 0.375 |
2.394 ± 0.198 | 2.716 ± 0.219 |
2.258 ± 0.185 | 6.927 ± 0.2 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.851 ± 0.127 | 2.666 ± 0.167 |
4.884 ± 0.187 | 5.059 ± 0.248 |
6.44 ± 0.382 | 5.806 ± 0.297 |
6.786 ± 0.325 | 7.691 ± 0.241 |
1.89 ± 0.111 | 2.739 ± 0.16 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
