
Methanomassiliicoccales archaeon RumEn M1
Taxonomy: cellular organisms; Archaea; Candidatus Thermoplasmatota; Thermoplasmata; Methanomassiliicoccales; unclassified Methanomassiliicoccales
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
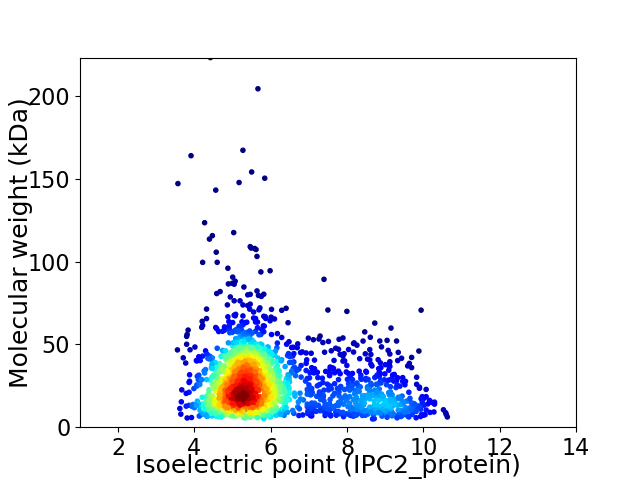
Virtual 2D-PAGE plot for 1877 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
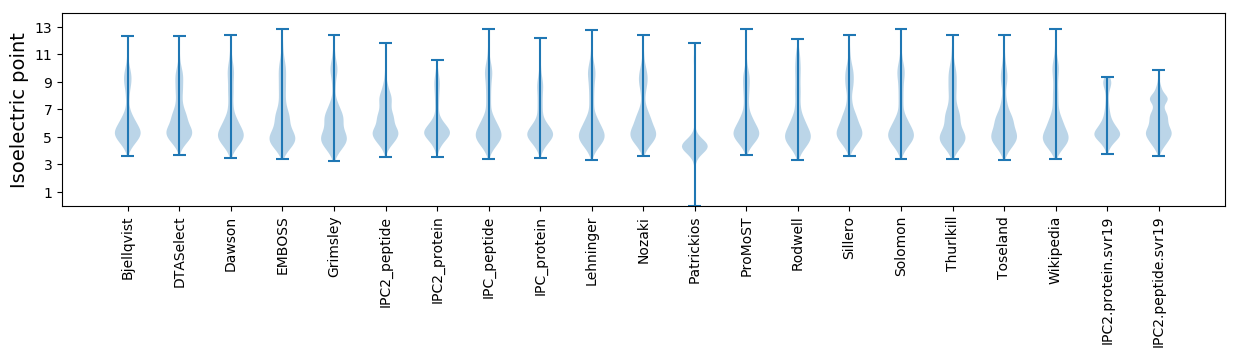
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q4BC58|A0A0Q4BC58_9ARCH B12-binding domain-containing protein OS=Methanomassiliicoccales archaeon RumEn M1 OX=1713724 GN=AOA80_07865 PE=4 SV=1
MM1 pKa = 7.18SAEE4 pKa = 4.38TGDD7 pKa = 3.68PWTDD11 pKa = 2.8IPEE14 pKa = 4.09NEE16 pKa = 4.18IIDD19 pKa = 3.91WGEE22 pKa = 3.39MWGTVIQFQYY32 pKa = 11.1VYY34 pKa = 10.39DD35 pKa = 4.09PEE37 pKa = 4.52VGNQAQTIEE46 pKa = 4.04WDD48 pKa = 3.79FGDD51 pKa = 4.26GSPRR55 pKa = 11.84STEE58 pKa = 3.46WNPRR62 pKa = 11.84HH63 pKa = 5.82TYY65 pKa = 9.97EE66 pKa = 4.52EE67 pKa = 4.6YY68 pKa = 9.05GTYY71 pKa = 9.99IVVQHH76 pKa = 5.19VTNTYY81 pKa = 10.6EE82 pKa = 4.63GFSEE86 pKa = 3.89DD87 pKa = 2.43WGYY90 pKa = 11.59YY91 pKa = 10.14RR92 pKa = 11.84MTIMGKK98 pKa = 9.13PYY100 pKa = 10.43VEE102 pKa = 4.13IVAPEE107 pKa = 4.4GAPQMEE113 pKa = 4.52KK114 pKa = 10.79VYY116 pKa = 9.26ATKK119 pKa = 9.36GTAPEE124 pKa = 4.26QPDD127 pKa = 3.42DD128 pKa = 5.04PIWKK132 pKa = 8.36GHH134 pKa = 5.53EE135 pKa = 3.85FLGYY139 pKa = 9.93YY140 pKa = 10.35ADD142 pKa = 5.0AEE144 pKa = 4.36FTVPFDD150 pKa = 3.08WTIPLDD156 pKa = 3.94RR157 pKa = 11.84AVTAYY162 pKa = 10.53AQFQNASVIPDD173 pKa = 4.5PDD175 pKa = 5.01DD176 pKa = 4.03EE177 pKa = 6.18DD178 pKa = 6.35PIDD181 pKa = 5.56DD182 pKa = 4.6EE183 pKa = 5.43DD184 pKa = 5.43PNGTDD189 pKa = 3.14SKK191 pKa = 11.9DD192 pKa = 3.48NITAGIGEE200 pKa = 4.28FFEE203 pKa = 4.83QYY205 pKa = 10.68GALTLGIAGGIVIAVFAIGVRR226 pKa = 11.84HH227 pKa = 5.61PVVLIVGIALLVIAALLKK245 pKa = 10.8FGVII249 pKa = 4.11
MM1 pKa = 7.18SAEE4 pKa = 4.38TGDD7 pKa = 3.68PWTDD11 pKa = 2.8IPEE14 pKa = 4.09NEE16 pKa = 4.18IIDD19 pKa = 3.91WGEE22 pKa = 3.39MWGTVIQFQYY32 pKa = 11.1VYY34 pKa = 10.39DD35 pKa = 4.09PEE37 pKa = 4.52VGNQAQTIEE46 pKa = 4.04WDD48 pKa = 3.79FGDD51 pKa = 4.26GSPRR55 pKa = 11.84STEE58 pKa = 3.46WNPRR62 pKa = 11.84HH63 pKa = 5.82TYY65 pKa = 9.97EE66 pKa = 4.52EE67 pKa = 4.6YY68 pKa = 9.05GTYY71 pKa = 9.99IVVQHH76 pKa = 5.19VTNTYY81 pKa = 10.6EE82 pKa = 4.63GFSEE86 pKa = 3.89DD87 pKa = 2.43WGYY90 pKa = 11.59YY91 pKa = 10.14RR92 pKa = 11.84MTIMGKK98 pKa = 9.13PYY100 pKa = 10.43VEE102 pKa = 4.13IVAPEE107 pKa = 4.4GAPQMEE113 pKa = 4.52KK114 pKa = 10.79VYY116 pKa = 9.26ATKK119 pKa = 9.36GTAPEE124 pKa = 4.26QPDD127 pKa = 3.42DD128 pKa = 5.04PIWKK132 pKa = 8.36GHH134 pKa = 5.53EE135 pKa = 3.85FLGYY139 pKa = 9.93YY140 pKa = 10.35ADD142 pKa = 5.0AEE144 pKa = 4.36FTVPFDD150 pKa = 3.08WTIPLDD156 pKa = 3.94RR157 pKa = 11.84AVTAYY162 pKa = 10.53AQFQNASVIPDD173 pKa = 4.5PDD175 pKa = 5.01DD176 pKa = 4.03EE177 pKa = 6.18DD178 pKa = 6.35PIDD181 pKa = 5.56DD182 pKa = 4.6EE183 pKa = 5.43DD184 pKa = 5.43PNGTDD189 pKa = 3.14SKK191 pKa = 11.9DD192 pKa = 3.48NITAGIGEE200 pKa = 4.28FFEE203 pKa = 4.83QYY205 pKa = 10.68GALTLGIAGGIVIAVFAIGVRR226 pKa = 11.84HH227 pKa = 5.61PVVLIVGIALLVIAALLKK245 pKa = 10.8FGVII249 pKa = 4.11
Molecular weight: 27.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q4BCL3|A0A0Q4BCL3_9ARCH Histidinol-phosphate aminotransferase OS=Methanomassiliicoccales archaeon RumEn M1 OX=1713724 GN=hisC PE=3 SV=1
MM1 pKa = 7.19KK2 pKa = 9.96HH3 pKa = 6.14YY4 pKa = 10.65EE5 pKa = 3.66IDD7 pKa = 3.6KK8 pKa = 9.34YY9 pKa = 11.28AHH11 pKa = 6.37SSSLFQFDD19 pKa = 3.86PRR21 pKa = 11.84VKK23 pKa = 9.79LACAISLIVVTAFLRR38 pKa = 11.84SMEE41 pKa = 3.82AVMVVLLFTALLVLFSRR58 pKa = 11.84VPLGHH63 pKa = 6.9LWKK66 pKa = 10.48NLALAVPFIVVPSLALLFTSGPYY89 pKa = 9.2NAAVLAMRR97 pKa = 11.84ILASVLALTAVISTTPLFDD116 pKa = 4.73LLRR119 pKa = 11.84TLRR122 pKa = 11.84WFRR125 pKa = 11.84MPKK128 pKa = 9.76LLSSIMMFAYY138 pKa = 10.16RR139 pKa = 11.84FIFVLIDD146 pKa = 3.02EE147 pKa = 4.62MDD149 pKa = 3.65RR150 pKa = 11.84MKK152 pKa = 9.9MARR155 pKa = 11.84MARR158 pKa = 11.84GYY160 pKa = 8.73TGRR163 pKa = 11.84GNILSRR169 pKa = 11.84DD170 pKa = 3.61VFRR173 pKa = 11.84TLSFTAGMIFVRR185 pKa = 11.84SNSRR189 pKa = 11.84ATRR192 pKa = 11.84IYY194 pKa = 10.81DD195 pKa = 3.4ALLARR200 pKa = 11.84GYY202 pKa = 10.73SGDD205 pKa = 3.25VHH207 pKa = 6.33TLDD210 pKa = 3.14RR211 pKa = 11.84MKK213 pKa = 10.82VRR215 pKa = 11.84GRR217 pKa = 11.84DD218 pKa = 3.55VPSAQCSSASEE229 pKa = 4.09RR230 pKa = 3.77
MM1 pKa = 7.19KK2 pKa = 9.96HH3 pKa = 6.14YY4 pKa = 10.65EE5 pKa = 3.66IDD7 pKa = 3.6KK8 pKa = 9.34YY9 pKa = 11.28AHH11 pKa = 6.37SSSLFQFDD19 pKa = 3.86PRR21 pKa = 11.84VKK23 pKa = 9.79LACAISLIVVTAFLRR38 pKa = 11.84SMEE41 pKa = 3.82AVMVVLLFTALLVLFSRR58 pKa = 11.84VPLGHH63 pKa = 6.9LWKK66 pKa = 10.48NLALAVPFIVVPSLALLFTSGPYY89 pKa = 9.2NAAVLAMRR97 pKa = 11.84ILASVLALTAVISTTPLFDD116 pKa = 4.73LLRR119 pKa = 11.84TLRR122 pKa = 11.84WFRR125 pKa = 11.84MPKK128 pKa = 9.76LLSSIMMFAYY138 pKa = 10.16RR139 pKa = 11.84FIFVLIDD146 pKa = 3.02EE147 pKa = 4.62MDD149 pKa = 3.65RR150 pKa = 11.84MKK152 pKa = 9.9MARR155 pKa = 11.84MARR158 pKa = 11.84GYY160 pKa = 8.73TGRR163 pKa = 11.84GNILSRR169 pKa = 11.84DD170 pKa = 3.61VFRR173 pKa = 11.84TLSFTAGMIFVRR185 pKa = 11.84SNSRR189 pKa = 11.84ATRR192 pKa = 11.84IYY194 pKa = 10.81DD195 pKa = 3.4ALLARR200 pKa = 11.84GYY202 pKa = 10.73SGDD205 pKa = 3.25VHH207 pKa = 6.33TLDD210 pKa = 3.14RR211 pKa = 11.84MKK213 pKa = 10.82VRR215 pKa = 11.84GRR217 pKa = 11.84DD218 pKa = 3.55VPSAQCSSASEE229 pKa = 4.09RR230 pKa = 3.77
Molecular weight: 25.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
479373 |
44 |
2088 |
255.4 |
28.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.212 ± 0.069 | 1.23 ± 0.028 |
5.977 ± 0.049 | 7.174 ± 0.064 |
3.447 ± 0.038 | 8.189 ± 0.06 |
1.906 ± 0.025 | 5.819 ± 0.051 |
4.073 ± 0.054 | 9.692 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.371 ± 0.03 | 2.755 ± 0.038 |
4.5 ± 0.034 | 2.522 ± 0.026 |
7.037 ± 0.077 | 6.099 ± 0.051 |
4.958 ± 0.047 | 8.214 ± 0.05 |
1.069 ± 0.021 | 2.757 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
