
Beihai tombus-like virus 11
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.71
Get precalculated fractions of proteins
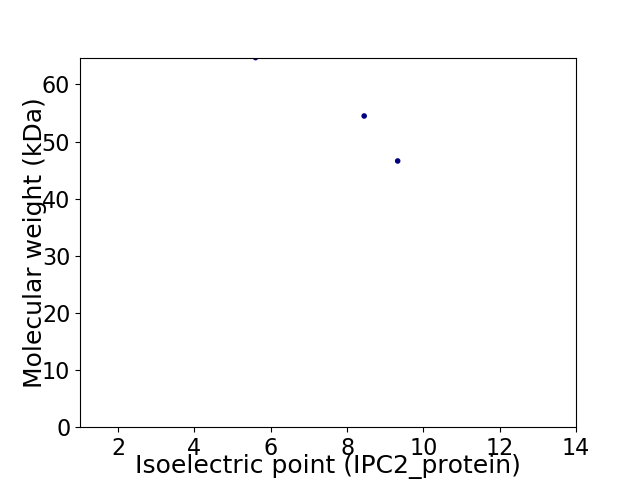
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFR2|A0A1L3KFR2_9VIRU RNA-directed RNA polymerase OS=Beihai tombus-like virus 11 OX=1922714 PE=4 SV=1
MM1 pKa = 7.49VSACQPCTDD10 pKa = 4.01NGAQYY15 pKa = 9.11VQEE18 pKa = 4.35VIAPSGATSCSGPPDD33 pKa = 3.88GCDD36 pKa = 3.1QSATKK41 pKa = 10.63GKK43 pKa = 8.32TRR45 pKa = 11.84DD46 pKa = 3.37QFVLSRR52 pKa = 11.84PDD54 pKa = 3.58FLPEE58 pKa = 3.82EE59 pKa = 4.37AWDD62 pKa = 3.65MVAFLTPYY70 pKa = 10.06YY71 pKa = 9.72VQQLVIVAFPSTVTPTQVRR90 pKa = 11.84TFLRR94 pKa = 11.84YY95 pKa = 9.03ILNSIPSEE103 pKa = 3.63EE104 pKa = 4.24WMGKK108 pKa = 7.55QLPGWHH114 pKa = 6.37NPADD118 pKa = 3.63YY119 pKa = 10.76SIYY122 pKa = 10.33DD123 pKa = 3.53DD124 pKa = 4.25NGNVVAGVPTMGVGWHH140 pKa = 6.5CPRR143 pKa = 11.84LLRR146 pKa = 11.84NLFPDD151 pKa = 4.02FGKK154 pKa = 10.26NPRR157 pKa = 11.84FVEE160 pKa = 4.24LVRR163 pKa = 11.84EE164 pKa = 4.04ARR166 pKa = 11.84RR167 pKa = 11.84WGRR170 pKa = 11.84YY171 pKa = 7.12LTITPVANATQFKK184 pKa = 10.43GRR186 pKa = 11.84IASASLQFNSALNPYY201 pKa = 9.73SLAITAGLSGPDD213 pKa = 4.13PLPPAISFPIGFVSLTLSLQLSRR236 pKa = 11.84DD237 pKa = 3.09RR238 pKa = 11.84LFYY241 pKa = 10.5YY242 pKa = 10.57DD243 pKa = 4.24GIRR246 pKa = 11.84WHH248 pKa = 6.69NIPWLNNPTEE258 pKa = 4.26VQFQSYY264 pKa = 10.49YY265 pKa = 11.14GFSIWNEE272 pKa = 3.79TGTDD276 pKa = 3.06ILVNSQAVITVRR288 pKa = 11.84ITITLTGVATVSWFVGPQLVHH309 pKa = 6.3QFTGDD314 pKa = 3.23VDD316 pKa = 3.47IGDD319 pKa = 4.29PLDD322 pKa = 3.57NQFLRR327 pKa = 11.84KK328 pKa = 9.45RR329 pKa = 11.84EE330 pKa = 3.79FDD332 pKa = 3.68SPSAAVSNEE341 pKa = 3.29LARR344 pKa = 11.84YY345 pKa = 8.94SVTPEE350 pKa = 3.75FSFEE354 pKa = 4.08ALLQADD360 pKa = 3.95EE361 pKa = 4.44KK362 pKa = 11.3SYY364 pKa = 10.17VNNWIEE370 pKa = 4.29GAHH373 pKa = 5.89SRR375 pKa = 11.84QAYY378 pKa = 9.71GKK380 pKa = 9.54DD381 pKa = 2.92HH382 pKa = 7.41LDD384 pKa = 3.51FVSTRR389 pKa = 11.84EE390 pKa = 3.85WRR392 pKa = 11.84PLIRR396 pKa = 11.84SANNVEE402 pKa = 4.4TVYY405 pKa = 11.32LDD407 pKa = 3.55ALSVKK412 pKa = 10.08RR413 pKa = 11.84DD414 pKa = 3.49LVDD417 pKa = 5.99LSGKK421 pKa = 7.21WQIDD425 pKa = 3.61YY426 pKa = 8.06ATNLDD431 pKa = 4.13PASSAAIVYY440 pKa = 7.04GTHH443 pKa = 5.88YY444 pKa = 10.08QFCCEE449 pKa = 3.98TSSEE453 pKa = 3.93FMLFKK458 pKa = 10.78RR459 pKa = 11.84EE460 pKa = 4.0VPEE463 pKa = 4.31KK464 pKa = 10.92DD465 pKa = 2.79EE466 pKa = 4.51GAIEE470 pKa = 4.03MAQVLQTALPHH481 pKa = 6.15TYY483 pKa = 9.04PAKK486 pKa = 10.69FNDD489 pKa = 3.97GGILPLILSKK499 pKa = 10.7VKK501 pKa = 10.49RR502 pKa = 11.84VGTNGLVGVAKK513 pKa = 10.78GIVNEE518 pKa = 3.91LLGGIRR524 pKa = 11.84EE525 pKa = 4.27IGTADD530 pKa = 4.06RR531 pKa = 11.84LIGYY535 pKa = 8.9PNMSTMYY542 pKa = 10.46GNNAVALYY550 pKa = 10.27NSNGNGNSNGNGNGKK565 pKa = 9.5KK566 pKa = 9.98KK567 pKa = 10.87KK568 pKa = 9.64NGNGNGVIPSIFSRR582 pKa = 11.84TMNII586 pKa = 3.26
MM1 pKa = 7.49VSACQPCTDD10 pKa = 4.01NGAQYY15 pKa = 9.11VQEE18 pKa = 4.35VIAPSGATSCSGPPDD33 pKa = 3.88GCDD36 pKa = 3.1QSATKK41 pKa = 10.63GKK43 pKa = 8.32TRR45 pKa = 11.84DD46 pKa = 3.37QFVLSRR52 pKa = 11.84PDD54 pKa = 3.58FLPEE58 pKa = 3.82EE59 pKa = 4.37AWDD62 pKa = 3.65MVAFLTPYY70 pKa = 10.06YY71 pKa = 9.72VQQLVIVAFPSTVTPTQVRR90 pKa = 11.84TFLRR94 pKa = 11.84YY95 pKa = 9.03ILNSIPSEE103 pKa = 3.63EE104 pKa = 4.24WMGKK108 pKa = 7.55QLPGWHH114 pKa = 6.37NPADD118 pKa = 3.63YY119 pKa = 10.76SIYY122 pKa = 10.33DD123 pKa = 3.53DD124 pKa = 4.25NGNVVAGVPTMGVGWHH140 pKa = 6.5CPRR143 pKa = 11.84LLRR146 pKa = 11.84NLFPDD151 pKa = 4.02FGKK154 pKa = 10.26NPRR157 pKa = 11.84FVEE160 pKa = 4.24LVRR163 pKa = 11.84EE164 pKa = 4.04ARR166 pKa = 11.84RR167 pKa = 11.84WGRR170 pKa = 11.84YY171 pKa = 7.12LTITPVANATQFKK184 pKa = 10.43GRR186 pKa = 11.84IASASLQFNSALNPYY201 pKa = 9.73SLAITAGLSGPDD213 pKa = 4.13PLPPAISFPIGFVSLTLSLQLSRR236 pKa = 11.84DD237 pKa = 3.09RR238 pKa = 11.84LFYY241 pKa = 10.5YY242 pKa = 10.57DD243 pKa = 4.24GIRR246 pKa = 11.84WHH248 pKa = 6.69NIPWLNNPTEE258 pKa = 4.26VQFQSYY264 pKa = 10.49YY265 pKa = 11.14GFSIWNEE272 pKa = 3.79TGTDD276 pKa = 3.06ILVNSQAVITVRR288 pKa = 11.84ITITLTGVATVSWFVGPQLVHH309 pKa = 6.3QFTGDD314 pKa = 3.23VDD316 pKa = 3.47IGDD319 pKa = 4.29PLDD322 pKa = 3.57NQFLRR327 pKa = 11.84KK328 pKa = 9.45RR329 pKa = 11.84EE330 pKa = 3.79FDD332 pKa = 3.68SPSAAVSNEE341 pKa = 3.29LARR344 pKa = 11.84YY345 pKa = 8.94SVTPEE350 pKa = 3.75FSFEE354 pKa = 4.08ALLQADD360 pKa = 3.95EE361 pKa = 4.44KK362 pKa = 11.3SYY364 pKa = 10.17VNNWIEE370 pKa = 4.29GAHH373 pKa = 5.89SRR375 pKa = 11.84QAYY378 pKa = 9.71GKK380 pKa = 9.54DD381 pKa = 2.92HH382 pKa = 7.41LDD384 pKa = 3.51FVSTRR389 pKa = 11.84EE390 pKa = 3.85WRR392 pKa = 11.84PLIRR396 pKa = 11.84SANNVEE402 pKa = 4.4TVYY405 pKa = 11.32LDD407 pKa = 3.55ALSVKK412 pKa = 10.08RR413 pKa = 11.84DD414 pKa = 3.49LVDD417 pKa = 5.99LSGKK421 pKa = 7.21WQIDD425 pKa = 3.61YY426 pKa = 8.06ATNLDD431 pKa = 4.13PASSAAIVYY440 pKa = 7.04GTHH443 pKa = 5.88YY444 pKa = 10.08QFCCEE449 pKa = 3.98TSSEE453 pKa = 3.93FMLFKK458 pKa = 10.78RR459 pKa = 11.84EE460 pKa = 4.0VPEE463 pKa = 4.31KK464 pKa = 10.92DD465 pKa = 2.79EE466 pKa = 4.51GAIEE470 pKa = 4.03MAQVLQTALPHH481 pKa = 6.15TYY483 pKa = 9.04PAKK486 pKa = 10.69FNDD489 pKa = 3.97GGILPLILSKK499 pKa = 10.7VKK501 pKa = 10.49RR502 pKa = 11.84VGTNGLVGVAKK513 pKa = 10.78GIVNEE518 pKa = 3.91LLGGIRR524 pKa = 11.84EE525 pKa = 4.27IGTADD530 pKa = 4.06RR531 pKa = 11.84LIGYY535 pKa = 8.9PNMSTMYY542 pKa = 10.46GNNAVALYY550 pKa = 10.27NSNGNGNSNGNGNGKK565 pKa = 9.5KK566 pKa = 9.98KK567 pKa = 10.87KK568 pKa = 9.64NGNGNGVIPSIFSRR582 pKa = 11.84TMNII586 pKa = 3.26
Molecular weight: 64.69 kDa
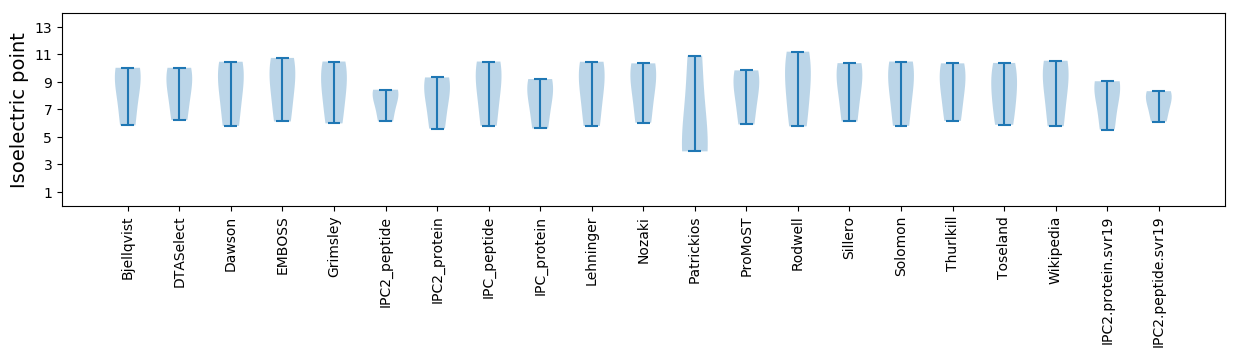
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFI3|A0A1L3KFI3_9VIRU Uncharacterized protein OS=Beihai tombus-like virus 11 OX=1922714 PE=4 SV=1
MM1 pKa = 7.22SRR3 pKa = 11.84SRR5 pKa = 11.84DD6 pKa = 3.59HH7 pKa = 6.78VPRR10 pKa = 11.84NLTEE14 pKa = 3.94VVEE17 pKa = 5.52FYY19 pKa = 10.79PHH21 pKa = 7.08LRR23 pKa = 11.84KK24 pKa = 9.58QVDD27 pKa = 3.18ALFRR31 pKa = 11.84KK32 pKa = 9.58ARR34 pKa = 11.84LADD37 pKa = 3.98GQPKK41 pKa = 9.65PKK43 pKa = 9.56PSKK46 pKa = 10.17KK47 pKa = 9.32PAPKK51 pKa = 9.97VVRR54 pKa = 11.84CLWCNSKK61 pKa = 10.56GYY63 pKa = 8.34RR64 pKa = 11.84TYY66 pKa = 11.66GDD68 pKa = 4.63DD69 pKa = 4.01DD70 pKa = 4.6LGYY73 pKa = 9.29TIADD77 pKa = 3.81CFRR80 pKa = 11.84CNTIFTFKK88 pKa = 10.55EE89 pKa = 3.75EE90 pKa = 3.86KK91 pKa = 11.03GEE93 pKa = 4.0IKK95 pKa = 10.44LRR97 pKa = 11.84SHH99 pKa = 7.35PFYY102 pKa = 10.94KK103 pKa = 10.25KK104 pKa = 10.12EE105 pKa = 4.11EE106 pKa = 4.11NVKK109 pKa = 10.44RR110 pKa = 11.84AVEE113 pKa = 4.22NIGKK117 pKa = 9.34EE118 pKa = 3.92VGNALGQIASKK129 pKa = 10.06SEE131 pKa = 4.01KK132 pKa = 8.48PNEE135 pKa = 3.74WVTVGKK141 pKa = 10.12NGKK144 pKa = 7.87PVKK147 pKa = 10.12KK148 pKa = 10.51DD149 pKa = 2.86ADD151 pKa = 3.79NKK153 pKa = 9.16VKK155 pKa = 10.53KK156 pKa = 9.96PKK158 pKa = 9.8VKK160 pKa = 10.57KK161 pKa = 9.88EE162 pKa = 3.62VSTKK166 pKa = 10.45VKK168 pKa = 10.45SPKK171 pKa = 9.72AKK173 pKa = 10.15KK174 pKa = 10.05DD175 pKa = 3.4VAVQASTPKK184 pKa = 10.29SKK186 pKa = 10.35KK187 pKa = 9.42GKK189 pKa = 8.47KK190 pKa = 9.75KK191 pKa = 10.14KK192 pKa = 10.54VKK194 pKa = 9.05TPKK197 pKa = 9.26GTNTPLIQRR206 pKa = 11.84KK207 pKa = 8.56AVTTDD212 pKa = 2.62MDD214 pKa = 3.69AVARR218 pKa = 11.84KK219 pKa = 9.61LKK221 pKa = 10.82AIAKK225 pKa = 9.25KK226 pKa = 10.48VNQEE230 pKa = 3.59RR231 pKa = 11.84SKK233 pKa = 10.79VQQSKK238 pKa = 10.33PKK240 pKa = 10.55AKK242 pKa = 7.65PTPPKK247 pKa = 10.38AVEE250 pKa = 4.11KK251 pKa = 9.64TVQEE255 pKa = 4.33PLPQRR260 pKa = 11.84VFPVVVQEE268 pKa = 4.07VLRR271 pKa = 11.84DD272 pKa = 3.81KK273 pKa = 10.69PIKK276 pKa = 9.95CQEE279 pKa = 3.89TEE281 pKa = 4.16EE282 pKa = 4.25IKK284 pKa = 11.0LVDD287 pKa = 3.8MPNINVDD294 pKa = 3.74RR295 pKa = 11.84MMKK298 pKa = 9.89TYY300 pKa = 8.31PTDD303 pKa = 3.36EE304 pKa = 5.15AEE306 pKa = 4.67FKK308 pKa = 11.24AMSANFRR315 pKa = 11.84QEE317 pKa = 4.63ALAQPAQAKK326 pKa = 9.19HH327 pKa = 4.72WLKK330 pKa = 10.82AADD333 pKa = 3.96RR334 pKa = 11.84LIYY337 pKa = 10.22SLPTRR342 pKa = 11.84VQVRR346 pKa = 11.84SPKK349 pKa = 10.6GLMRR353 pKa = 11.84AKK355 pKa = 10.23RR356 pKa = 11.84DD357 pKa = 3.92LITSRR362 pKa = 11.84PYY364 pKa = 8.86MHH366 pKa = 7.69GYY368 pKa = 9.2LQLCNQLDD376 pKa = 4.18DD377 pKa = 5.6FEE379 pKa = 4.94MKK381 pKa = 10.41HH382 pKa = 6.71SEE384 pKa = 4.12EE385 pKa = 5.58DD386 pKa = 3.55FMQAAAMQVALRR398 pKa = 11.84PLKK401 pKa = 10.64KK402 pKa = 10.08SWSKK406 pKa = 10.91SKK408 pKa = 10.69KK409 pKa = 9.54
MM1 pKa = 7.22SRR3 pKa = 11.84SRR5 pKa = 11.84DD6 pKa = 3.59HH7 pKa = 6.78VPRR10 pKa = 11.84NLTEE14 pKa = 3.94VVEE17 pKa = 5.52FYY19 pKa = 10.79PHH21 pKa = 7.08LRR23 pKa = 11.84KK24 pKa = 9.58QVDD27 pKa = 3.18ALFRR31 pKa = 11.84KK32 pKa = 9.58ARR34 pKa = 11.84LADD37 pKa = 3.98GQPKK41 pKa = 9.65PKK43 pKa = 9.56PSKK46 pKa = 10.17KK47 pKa = 9.32PAPKK51 pKa = 9.97VVRR54 pKa = 11.84CLWCNSKK61 pKa = 10.56GYY63 pKa = 8.34RR64 pKa = 11.84TYY66 pKa = 11.66GDD68 pKa = 4.63DD69 pKa = 4.01DD70 pKa = 4.6LGYY73 pKa = 9.29TIADD77 pKa = 3.81CFRR80 pKa = 11.84CNTIFTFKK88 pKa = 10.55EE89 pKa = 3.75EE90 pKa = 3.86KK91 pKa = 11.03GEE93 pKa = 4.0IKK95 pKa = 10.44LRR97 pKa = 11.84SHH99 pKa = 7.35PFYY102 pKa = 10.94KK103 pKa = 10.25KK104 pKa = 10.12EE105 pKa = 4.11EE106 pKa = 4.11NVKK109 pKa = 10.44RR110 pKa = 11.84AVEE113 pKa = 4.22NIGKK117 pKa = 9.34EE118 pKa = 3.92VGNALGQIASKK129 pKa = 10.06SEE131 pKa = 4.01KK132 pKa = 8.48PNEE135 pKa = 3.74WVTVGKK141 pKa = 10.12NGKK144 pKa = 7.87PVKK147 pKa = 10.12KK148 pKa = 10.51DD149 pKa = 2.86ADD151 pKa = 3.79NKK153 pKa = 9.16VKK155 pKa = 10.53KK156 pKa = 9.96PKK158 pKa = 9.8VKK160 pKa = 10.57KK161 pKa = 9.88EE162 pKa = 3.62VSTKK166 pKa = 10.45VKK168 pKa = 10.45SPKK171 pKa = 9.72AKK173 pKa = 10.15KK174 pKa = 10.05DD175 pKa = 3.4VAVQASTPKK184 pKa = 10.29SKK186 pKa = 10.35KK187 pKa = 9.42GKK189 pKa = 8.47KK190 pKa = 9.75KK191 pKa = 10.14KK192 pKa = 10.54VKK194 pKa = 9.05TPKK197 pKa = 9.26GTNTPLIQRR206 pKa = 11.84KK207 pKa = 8.56AVTTDD212 pKa = 2.62MDD214 pKa = 3.69AVARR218 pKa = 11.84KK219 pKa = 9.61LKK221 pKa = 10.82AIAKK225 pKa = 9.25KK226 pKa = 10.48VNQEE230 pKa = 3.59RR231 pKa = 11.84SKK233 pKa = 10.79VQQSKK238 pKa = 10.33PKK240 pKa = 10.55AKK242 pKa = 7.65PTPPKK247 pKa = 10.38AVEE250 pKa = 4.11KK251 pKa = 9.64TVQEE255 pKa = 4.33PLPQRR260 pKa = 11.84VFPVVVQEE268 pKa = 4.07VLRR271 pKa = 11.84DD272 pKa = 3.81KK273 pKa = 10.69PIKK276 pKa = 9.95CQEE279 pKa = 3.89TEE281 pKa = 4.16EE282 pKa = 4.25IKK284 pKa = 11.0LVDD287 pKa = 3.8MPNINVDD294 pKa = 3.74RR295 pKa = 11.84MMKK298 pKa = 9.89TYY300 pKa = 8.31PTDD303 pKa = 3.36EE304 pKa = 5.15AEE306 pKa = 4.67FKK308 pKa = 11.24AMSANFRR315 pKa = 11.84QEE317 pKa = 4.63ALAQPAQAKK326 pKa = 9.19HH327 pKa = 4.72WLKK330 pKa = 10.82AADD333 pKa = 3.96RR334 pKa = 11.84LIYY337 pKa = 10.22SLPTRR342 pKa = 11.84VQVRR346 pKa = 11.84SPKK349 pKa = 10.6GLMRR353 pKa = 11.84AKK355 pKa = 10.23RR356 pKa = 11.84DD357 pKa = 3.92LITSRR362 pKa = 11.84PYY364 pKa = 8.86MHH366 pKa = 7.69GYY368 pKa = 9.2LQLCNQLDD376 pKa = 4.18DD377 pKa = 5.6FEE379 pKa = 4.94MKK381 pKa = 10.41HH382 pKa = 6.71SEE384 pKa = 4.12EE385 pKa = 5.58DD386 pKa = 3.55FMQAAAMQVALRR398 pKa = 11.84PLKK401 pKa = 10.64KK402 pKa = 10.08SWSKK406 pKa = 10.91SKK408 pKa = 10.69KK409 pKa = 9.54
Molecular weight: 46.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1466 |
409 |
586 |
488.7 |
55.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.094 ± 0.405 | 1.91 ± 0.512 |
5.184 ± 0.049 | 5.116 ± 0.442 |
3.752 ± 0.454 | 5.798 ± 1.197 |
1.842 ± 0.365 | 5.252 ± 0.859 |
8.254 ± 2.994 | 8.117 ± 0.825 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.456 ± 0.478 | 5.116 ± 0.764 |
6.003 ± 0.63 | 3.888 ± 0.463 |
6.003 ± 0.573 | 6.958 ± 0.624 |
4.638 ± 0.9 | 7.094 ± 0.793 |
1.501 ± 0.272 | 4.025 ± 0.747 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
