
Salipiger sp. IMCC34102
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Salipiger; unclassified Salipiger
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
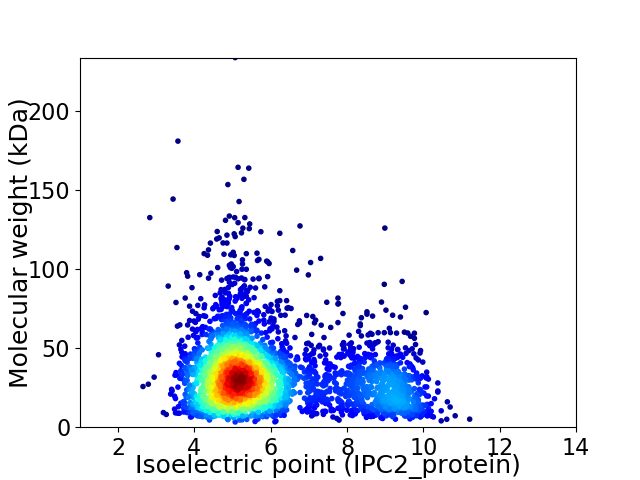
Virtual 2D-PAGE plot for 3341 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
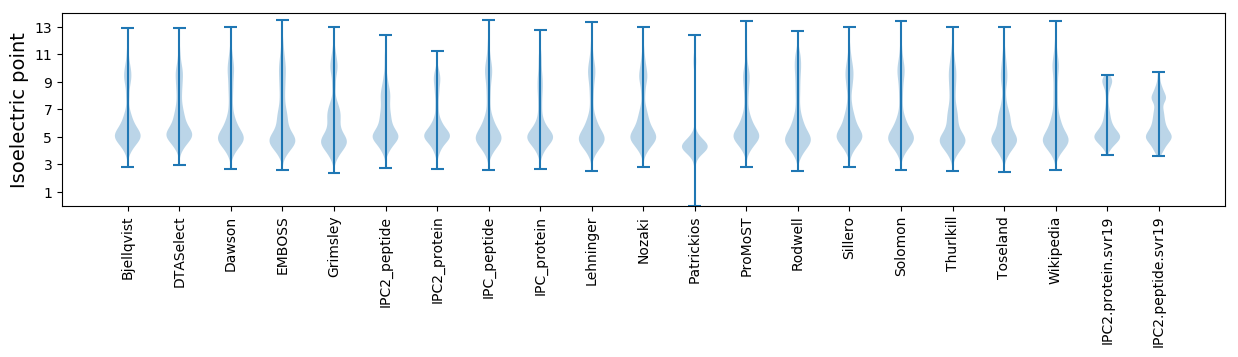
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q3Z7T4|A0A4Q3Z7T4_9RHOB Leucyl aminopeptidase family protein OS=Salipiger sp. IMCC34102 OX=2510647 GN=EU805_00405 PE=3 SV=1
MM1 pKa = 7.42NKK3 pKa = 9.63FLAIATASVVASAAQAQEE21 pKa = 4.15TVVWWDD27 pKa = 3.94FLAGGDD33 pKa = 4.04GVRR36 pKa = 11.84MKK38 pKa = 11.15QMIEE42 pKa = 4.21DD43 pKa = 4.05FNAAHH48 pKa = 6.82EE49 pKa = 4.48GEE51 pKa = 4.26IVIDD55 pKa = 3.63ATTLEE60 pKa = 4.33WGVPYY65 pKa = 8.75YY66 pKa = 10.73TKK68 pKa = 10.58VQTSVAVGEE77 pKa = 4.86APDD80 pKa = 2.99IMTYY84 pKa = 9.79HH85 pKa = 7.42SSRR88 pKa = 11.84IPLAIDD94 pKa = 2.99QGLLQEE100 pKa = 4.58LTPADD105 pKa = 4.5FEE107 pKa = 4.92TMGLGEE113 pKa = 5.67DD114 pKa = 4.59DD115 pKa = 4.45FAPSIWDD122 pKa = 3.39AVSADD127 pKa = 3.19GAQYY131 pKa = 11.34GMPLDD136 pKa = 3.43THH138 pKa = 6.95PIVLYY143 pKa = 9.49YY144 pKa = 10.97NKK146 pKa = 10.27DD147 pKa = 3.35ALEE150 pKa = 4.07EE151 pKa = 4.13AGRR154 pKa = 11.84LTEE157 pKa = 5.66DD158 pKa = 4.09GLPMGLDD165 pKa = 3.17SRR167 pKa = 11.84EE168 pKa = 4.22GFTQTLQALQDD179 pKa = 3.72SGSVNFPLASVTADD193 pKa = 3.09GNFMFRR199 pKa = 11.84TIYY202 pKa = 11.0SLMGQQGGEE211 pKa = 4.07LMTDD215 pKa = 3.82GEE217 pKa = 5.01FLAGDD222 pKa = 3.67NPQKK226 pKa = 10.87LEE228 pKa = 3.85NALTVLQEE236 pKa = 3.61WTNAGFQSTYY246 pKa = 8.47TDD248 pKa = 3.59YY249 pKa = 10.57PATVALFTNGEE260 pKa = 4.13AGMMINGVWEE270 pKa = 4.56VPTMIDD276 pKa = 5.07LEE278 pKa = 4.55AEE280 pKa = 4.13GNLFNWGAVEE290 pKa = 4.27LPVIFDD296 pKa = 3.57QPATYY301 pKa = 10.52SDD303 pKa = 3.5SHH305 pKa = 7.55VFAIPAGNDD314 pKa = 2.86MSEE317 pKa = 4.37EE318 pKa = 3.59KK319 pKa = 10.26RR320 pKa = 11.84AAVLEE325 pKa = 4.3VMSWFSNNSLFWATAGHH342 pKa = 6.55IPANNSVQGTDD353 pKa = 3.58EE354 pKa = 4.2YY355 pKa = 11.62QAMEE359 pKa = 4.35PNATYY364 pKa = 10.99SSLTEE369 pKa = 3.85NMIYY373 pKa = 10.34DD374 pKa = 4.68PKK376 pKa = 10.81TPLAGVAGPIFDD388 pKa = 3.8VMSTYY393 pKa = 10.27FVPTLNGEE401 pKa = 4.17MDD403 pKa = 3.6PSEE406 pKa = 4.14AVEE409 pKa = 4.4EE410 pKa = 4.1ITYY413 pKa = 9.97EE414 pKa = 4.08LNDD417 pKa = 3.42MM418 pKa = 4.77
MM1 pKa = 7.42NKK3 pKa = 9.63FLAIATASVVASAAQAQEE21 pKa = 4.15TVVWWDD27 pKa = 3.94FLAGGDD33 pKa = 4.04GVRR36 pKa = 11.84MKK38 pKa = 11.15QMIEE42 pKa = 4.21DD43 pKa = 4.05FNAAHH48 pKa = 6.82EE49 pKa = 4.48GEE51 pKa = 4.26IVIDD55 pKa = 3.63ATTLEE60 pKa = 4.33WGVPYY65 pKa = 8.75YY66 pKa = 10.73TKK68 pKa = 10.58VQTSVAVGEE77 pKa = 4.86APDD80 pKa = 2.99IMTYY84 pKa = 9.79HH85 pKa = 7.42SSRR88 pKa = 11.84IPLAIDD94 pKa = 2.99QGLLQEE100 pKa = 4.58LTPADD105 pKa = 4.5FEE107 pKa = 4.92TMGLGEE113 pKa = 5.67DD114 pKa = 4.59DD115 pKa = 4.45FAPSIWDD122 pKa = 3.39AVSADD127 pKa = 3.19GAQYY131 pKa = 11.34GMPLDD136 pKa = 3.43THH138 pKa = 6.95PIVLYY143 pKa = 9.49YY144 pKa = 10.97NKK146 pKa = 10.27DD147 pKa = 3.35ALEE150 pKa = 4.07EE151 pKa = 4.13AGRR154 pKa = 11.84LTEE157 pKa = 5.66DD158 pKa = 4.09GLPMGLDD165 pKa = 3.17SRR167 pKa = 11.84EE168 pKa = 4.22GFTQTLQALQDD179 pKa = 3.72SGSVNFPLASVTADD193 pKa = 3.09GNFMFRR199 pKa = 11.84TIYY202 pKa = 11.0SLMGQQGGEE211 pKa = 4.07LMTDD215 pKa = 3.82GEE217 pKa = 5.01FLAGDD222 pKa = 3.67NPQKK226 pKa = 10.87LEE228 pKa = 3.85NALTVLQEE236 pKa = 3.61WTNAGFQSTYY246 pKa = 8.47TDD248 pKa = 3.59YY249 pKa = 10.57PATVALFTNGEE260 pKa = 4.13AGMMINGVWEE270 pKa = 4.56VPTMIDD276 pKa = 5.07LEE278 pKa = 4.55AEE280 pKa = 4.13GNLFNWGAVEE290 pKa = 4.27LPVIFDD296 pKa = 3.57QPATYY301 pKa = 10.52SDD303 pKa = 3.5SHH305 pKa = 7.55VFAIPAGNDD314 pKa = 2.86MSEE317 pKa = 4.37EE318 pKa = 3.59KK319 pKa = 10.26RR320 pKa = 11.84AAVLEE325 pKa = 4.3VMSWFSNNSLFWATAGHH342 pKa = 6.55IPANNSVQGTDD353 pKa = 3.58EE354 pKa = 4.2YY355 pKa = 11.62QAMEE359 pKa = 4.35PNATYY364 pKa = 10.99SSLTEE369 pKa = 3.85NMIYY373 pKa = 10.34DD374 pKa = 4.68PKK376 pKa = 10.81TPLAGVAGPIFDD388 pKa = 3.8VMSTYY393 pKa = 10.27FVPTLNGEE401 pKa = 4.17MDD403 pKa = 3.6PSEE406 pKa = 4.14AVEE409 pKa = 4.4EE410 pKa = 4.1ITYY413 pKa = 9.97EE414 pKa = 4.08LNDD417 pKa = 3.42MM418 pKa = 4.77
Molecular weight: 45.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q3YZ16|A0A4Q3YZ16_9RHOB Gfo/Idh/MocA family oxidoreductase OS=Salipiger sp. IMCC34102 OX=2510647 GN=EU805_03875 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.81IINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.31GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.81IINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.31GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1060431 |
28 |
2168 |
317.4 |
34.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.713 ± 0.052 | 0.859 ± 0.015 |
6.643 ± 0.042 | 5.712 ± 0.041 |
3.575 ± 0.026 | 8.799 ± 0.046 |
1.983 ± 0.021 | 4.934 ± 0.028 |
2.602 ± 0.033 | 9.992 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.664 ± 0.022 | 2.349 ± 0.021 |
5.243 ± 0.036 | 3.144 ± 0.021 |
7.145 ± 0.041 | 4.928 ± 0.036 |
5.853 ± 0.027 | 7.387 ± 0.036 |
1.374 ± 0.021 | 2.102 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
