
Nocardioides sp. PD653
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides; unclassified Nocardioides
Average proteome isoelectric point is 5.91
Get precalculated fractions of proteins
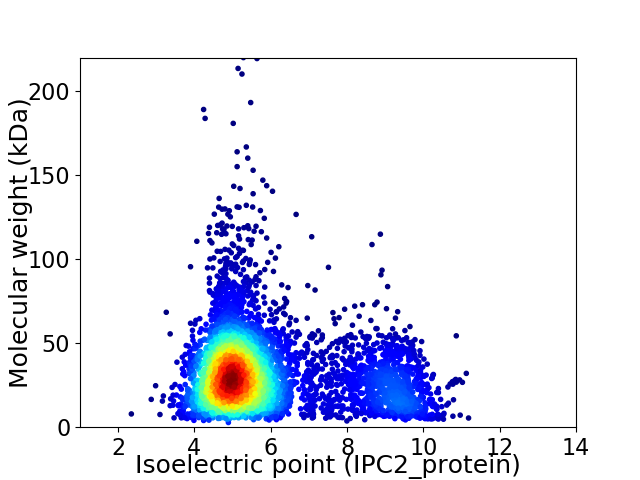
Virtual 2D-PAGE plot for 5032 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V1VZS1|A0A1V1VZS1_9ACTN Uncharacterized protein OS=Nocardioides sp. PD653 OX=393303 GN=PD653_1060 PE=4 SV=1
MM1 pKa = 7.32LTRR4 pKa = 11.84SRR6 pKa = 11.84AAFAGLAVVAAFTLAGCGGDD26 pKa = 3.55SDD28 pKa = 5.74SDD30 pKa = 4.04VATDD34 pKa = 3.93TPSASPTGPACDD46 pKa = 4.18YY47 pKa = 10.88PADD50 pKa = 3.81AQGAAKK56 pKa = 9.96KK57 pKa = 10.26VNPPPARR64 pKa = 11.84AAVSGEE70 pKa = 3.79VGATMKK76 pKa = 10.46TSAGDD81 pKa = 3.0IGLTLDD87 pKa = 5.25ADD89 pKa = 4.15TTPCTVNSFVSLAEE103 pKa = 3.56QGYY106 pKa = 10.27FDD108 pKa = 3.83GTTCHH113 pKa = 7.23RR114 pKa = 11.84LTTTDD119 pKa = 2.73AGIFVLQCGDD129 pKa = 3.46PTGTGMGGPGYY140 pKa = 10.79SFDD143 pKa = 5.15DD144 pKa = 3.89EE145 pKa = 5.04LSGSEE150 pKa = 4.29TYY152 pKa = 10.07PAGTLAMANAGPDD165 pKa = 3.6TNGSQFFLVYY175 pKa = 10.82ADD177 pKa = 3.92TPLPASYY184 pKa = 9.55TVFGTVDD191 pKa = 3.4DD192 pKa = 5.17AGLEE196 pKa = 4.18VLKK199 pKa = 11.16GIADD203 pKa = 3.83EE204 pKa = 4.62GTADD208 pKa = 4.48GGPDD212 pKa = 3.84GPPRR216 pKa = 11.84TPVDD220 pKa = 3.41IEE222 pKa = 4.6SVTVDD227 pKa = 2.93
MM1 pKa = 7.32LTRR4 pKa = 11.84SRR6 pKa = 11.84AAFAGLAVVAAFTLAGCGGDD26 pKa = 3.55SDD28 pKa = 5.74SDD30 pKa = 4.04VATDD34 pKa = 3.93TPSASPTGPACDD46 pKa = 4.18YY47 pKa = 10.88PADD50 pKa = 3.81AQGAAKK56 pKa = 9.96KK57 pKa = 10.26VNPPPARR64 pKa = 11.84AAVSGEE70 pKa = 3.79VGATMKK76 pKa = 10.46TSAGDD81 pKa = 3.0IGLTLDD87 pKa = 5.25ADD89 pKa = 4.15TTPCTVNSFVSLAEE103 pKa = 3.56QGYY106 pKa = 10.27FDD108 pKa = 3.83GTTCHH113 pKa = 7.23RR114 pKa = 11.84LTTTDD119 pKa = 2.73AGIFVLQCGDD129 pKa = 3.46PTGTGMGGPGYY140 pKa = 10.79SFDD143 pKa = 5.15DD144 pKa = 3.89EE145 pKa = 5.04LSGSEE150 pKa = 4.29TYY152 pKa = 10.07PAGTLAMANAGPDD165 pKa = 3.6TNGSQFFLVYY175 pKa = 10.82ADD177 pKa = 3.92TPLPASYY184 pKa = 9.55TVFGTVDD191 pKa = 3.4DD192 pKa = 5.17AGLEE196 pKa = 4.18VLKK199 pKa = 11.16GIADD203 pKa = 3.83EE204 pKa = 4.62GTADD208 pKa = 4.48GGPDD212 pKa = 3.84GPPRR216 pKa = 11.84TPVDD220 pKa = 3.41IEE222 pKa = 4.6SVTVDD227 pKa = 2.93
Molecular weight: 22.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V1WB83|A0A1V1WB83_9ACTN 3-oxoacyl-ACP reductase OS=Nocardioides sp. PD653 OX=393303 GN=PD653_4866 PE=4 SV=1
RR1 pKa = 7.53EE2 pKa = 4.15GHH4 pKa = 6.16PVAPVRR10 pKa = 11.84GGRR13 pKa = 11.84VRR15 pKa = 11.84GHH17 pKa = 5.84PRR19 pKa = 11.84RR20 pKa = 11.84PRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84GTLLRR29 pKa = 11.84PAGAGGRR36 pKa = 11.84GSPGAVRR43 pKa = 11.84PHH45 pKa = 7.28RR46 pKa = 11.84SRR48 pKa = 11.84SPPAARR54 pKa = 11.84RR55 pKa = 11.84GRR57 pKa = 11.84PVLRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84HH64 pKa = 5.78HH65 pKa = 6.5PGLVQRR71 pKa = 11.84HH72 pKa = 4.88RR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84VRR77 pKa = 11.84HH78 pKa = 5.55PRR80 pKa = 11.84PGRR83 pKa = 11.84RR84 pKa = 11.84PRR86 pKa = 11.84PRR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84HH91 pKa = 4.53HH92 pKa = 6.58RR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84PTAGRR100 pKa = 11.84PGVRR104 pKa = 11.84RR105 pKa = 11.84LVGRR109 pKa = 11.84PPVHH113 pKa = 5.9GRR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84PRR119 pKa = 11.84QEE121 pKa = 3.26PRR123 pKa = 11.84VFGVPRR129 pKa = 11.84RR130 pKa = 11.84PAPPGRR136 pKa = 11.84AQRR139 pKa = 11.84PFGRR143 pKa = 11.84RR144 pKa = 11.84TLRR147 pKa = 11.84HH148 pKa = 5.73AGEE151 pKa = 4.6HH152 pKa = 5.36EE153 pKa = 4.76VVPLPRR159 pKa = 11.84PGPRR163 pKa = 11.84LVPRR167 pKa = 11.84LVRR170 pKa = 11.84ARR172 pKa = 11.84ASGHH176 pKa = 5.68RR177 pKa = 11.84PQRR180 pKa = 11.84TPLEE184 pKa = 4.54PGPRR188 pKa = 11.84PHH190 pKa = 6.65PTAARR195 pKa = 11.84TGEE198 pKa = 4.03PHH200 pKa = 6.85PPHH203 pKa = 6.59LRR205 pKa = 11.84VPALHH210 pKa = 6.49PTIQVLRR217 pKa = 11.84RR218 pKa = 11.84RR219 pKa = 11.84PHH221 pKa = 5.32HH222 pKa = 6.36RR223 pKa = 11.84LRR225 pKa = 11.84PEE227 pKa = 3.81RR228 pKa = 11.84THH230 pKa = 7.09LLLQPRR236 pKa = 11.84PPVPPPPPAQNPRR249 pKa = 11.84QLVVRR254 pKa = 11.84TGRR257 pKa = 11.84TPGIPLAKK265 pKa = 9.55PAGALLPPRR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84PPRR279 pKa = 11.84YY280 pKa = 8.98VV281 pKa = 2.85
RR1 pKa = 7.53EE2 pKa = 4.15GHH4 pKa = 6.16PVAPVRR10 pKa = 11.84GGRR13 pKa = 11.84VRR15 pKa = 11.84GHH17 pKa = 5.84PRR19 pKa = 11.84RR20 pKa = 11.84PRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84GTLLRR29 pKa = 11.84PAGAGGRR36 pKa = 11.84GSPGAVRR43 pKa = 11.84PHH45 pKa = 7.28RR46 pKa = 11.84SRR48 pKa = 11.84SPPAARR54 pKa = 11.84RR55 pKa = 11.84GRR57 pKa = 11.84PVLRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84HH64 pKa = 5.78HH65 pKa = 6.5PGLVQRR71 pKa = 11.84HH72 pKa = 4.88RR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84VRR77 pKa = 11.84HH78 pKa = 5.55PRR80 pKa = 11.84PGRR83 pKa = 11.84RR84 pKa = 11.84PRR86 pKa = 11.84PRR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84HH91 pKa = 4.53HH92 pKa = 6.58RR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84PTAGRR100 pKa = 11.84PGVRR104 pKa = 11.84RR105 pKa = 11.84LVGRR109 pKa = 11.84PPVHH113 pKa = 5.9GRR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84PRR119 pKa = 11.84QEE121 pKa = 3.26PRR123 pKa = 11.84VFGVPRR129 pKa = 11.84RR130 pKa = 11.84PAPPGRR136 pKa = 11.84AQRR139 pKa = 11.84PFGRR143 pKa = 11.84RR144 pKa = 11.84TLRR147 pKa = 11.84HH148 pKa = 5.73AGEE151 pKa = 4.6HH152 pKa = 5.36EE153 pKa = 4.76VVPLPRR159 pKa = 11.84PGPRR163 pKa = 11.84LVPRR167 pKa = 11.84LVRR170 pKa = 11.84ARR172 pKa = 11.84ASGHH176 pKa = 5.68RR177 pKa = 11.84PQRR180 pKa = 11.84TPLEE184 pKa = 4.54PGPRR188 pKa = 11.84PHH190 pKa = 6.65PTAARR195 pKa = 11.84TGEE198 pKa = 4.03PHH200 pKa = 6.85PPHH203 pKa = 6.59LRR205 pKa = 11.84VPALHH210 pKa = 6.49PTIQVLRR217 pKa = 11.84RR218 pKa = 11.84RR219 pKa = 11.84PHH221 pKa = 5.32HH222 pKa = 6.36RR223 pKa = 11.84LRR225 pKa = 11.84PEE227 pKa = 3.81RR228 pKa = 11.84THH230 pKa = 7.09LLLQPRR236 pKa = 11.84PPVPPPPPAQNPRR249 pKa = 11.84QLVVRR254 pKa = 11.84TGRR257 pKa = 11.84TPGIPLAKK265 pKa = 9.55PAGALLPPRR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84PPRR279 pKa = 11.84YY280 pKa = 8.98VV281 pKa = 2.85
Molecular weight: 32.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1565263 |
27 |
2022 |
311.1 |
33.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.818 ± 0.043 | 0.754 ± 0.01 |
6.664 ± 0.029 | 5.634 ± 0.031 |
2.797 ± 0.019 | 9.12 ± 0.033 |
2.289 ± 0.018 | 3.627 ± 0.025 |
2.019 ± 0.024 | 10.099 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.897 ± 0.013 | 1.814 ± 0.018 |
5.675 ± 0.029 | 2.823 ± 0.017 |
7.492 ± 0.04 | 5.393 ± 0.024 |
6.228 ± 0.029 | 9.313 ± 0.032 |
1.531 ± 0.015 | 2.014 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
