
Bradyrhizobiaceae bacterium SG-6C
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Bradyrhizobiaceae; unclassified Bradyrhizobiaceae
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
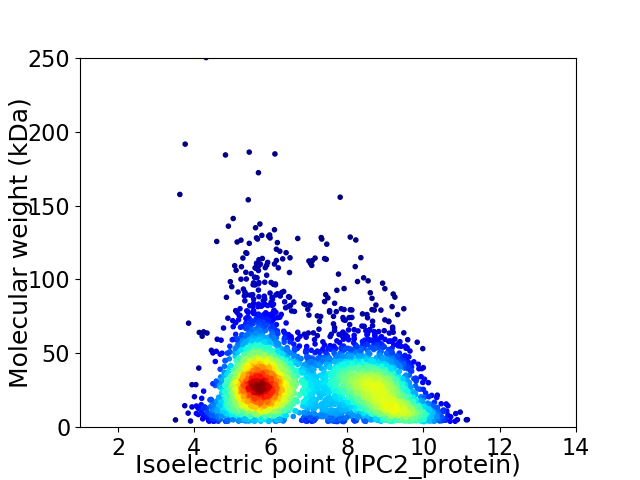
Virtual 2D-PAGE plot for 4236 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F7QN18|F7QN18_9BRAD DD-transpeptidase OS=Bradyrhizobiaceae bacterium SG-6C OX=709797 GN=CSIRO_3170 PE=3 SV=1
MM1 pKa = 7.45SATAAAGVLSNDD13 pKa = 3.08TFGADD18 pKa = 3.3GASVVGVAKK27 pKa = 9.44GTTNANLDD35 pKa = 3.55NTGTLGAGLHH45 pKa = 5.32GTYY48 pKa = 8.88GTLTLNADD56 pKa = 3.45GSYY59 pKa = 10.82SYY61 pKa = 11.01IRR63 pKa = 11.84DD64 pKa = 3.61ANSAGGVTDD73 pKa = 3.91TFTYY77 pKa = 9.47TIKK80 pKa = 11.15DD81 pKa = 3.23GDD83 pKa = 4.1GDD85 pKa = 4.18LSHH88 pKa = 6.71TTLTITLPDD97 pKa = 3.59ATPSYY102 pKa = 9.62TLPTAGNGVTTVYY115 pKa = 10.43EE116 pKa = 3.86AGLAVRR122 pKa = 11.84SGEE125 pKa = 4.0PAGSHH130 pKa = 6.16IGATNITATGVIGVSSVDD148 pKa = 3.52GVGSVSLNGQTVTGTSAGTATTVLSNSTGTLTAWYY183 pKa = 7.85EE184 pKa = 3.87AATSQIHH191 pKa = 4.33YY192 pKa = 9.06TYY194 pKa = 10.71TLLDD198 pKa = 3.46NTLNTSGTSISVPVGVTDD216 pKa = 5.78LDD218 pKa = 3.99GDD220 pKa = 4.22TPASGGNLVITIVDD234 pKa = 3.92DD235 pKa = 4.4APVAVSDD242 pKa = 4.05TNSLGTASVGSQASGNLIAGTITSGSGTGGADD274 pKa = 3.23TQGADD279 pKa = 3.41GAHH282 pKa = 6.65IGSVTGGASDD292 pKa = 3.64GSGGFNVAGTYY303 pKa = 7.48GTLHH307 pKa = 6.21VDD309 pKa = 3.26GNGAYY314 pKa = 10.22SYY316 pKa = 10.46TRR318 pKa = 11.84TSAAGGTDD326 pKa = 3.23TFHH329 pKa = 6.37YY330 pKa = 9.68TLTDD334 pKa = 3.38GDD336 pKa = 4.4GDD338 pKa = 3.58ISNQADD344 pKa = 3.77LTITLDD350 pKa = 3.24DD351 pKa = 3.65TGRR354 pKa = 11.84LFVGSNDD361 pKa = 4.2SDD363 pKa = 4.96DD364 pKa = 4.47NPNDD368 pKa = 3.47PAHH371 pKa = 6.22FVDD374 pKa = 4.2VSSNVTGVINGGGGNDD390 pKa = 4.39TIAGDD395 pKa = 3.96PGATPTISAGSSANIVLVLDD415 pKa = 4.2TTASLTAQQMSAMQQAVGALLNSLYY440 pKa = 10.96NSGAANVRR448 pKa = 11.84VSMYY452 pKa = 10.83AFGGGYY458 pKa = 7.04TQLGTYY464 pKa = 10.27NIVANSHH471 pKa = 6.01SNAAADD477 pKa = 3.62AAALVSALDD486 pKa = 3.65AANADD491 pKa = 3.71GSITNSGSRR500 pKa = 11.84LYY502 pKa = 10.38IGADD506 pKa = 3.15PGSNTSYY513 pKa = 11.62ADD515 pKa = 3.47GLDD518 pKa = 3.4AVIDD522 pKa = 3.92YY523 pKa = 7.37LTPGTSGGDD532 pKa = 3.43PVSAVPLTGASVNKK546 pKa = 10.15VIFISDD552 pKa = 4.2GEE554 pKa = 4.36PTTNATQTQINTITGTGAGQYY575 pKa = 10.1GYY577 pKa = 9.0TIEE580 pKa = 4.87AVGLNVAGSPNAVAALNALDD600 pKa = 4.64SGHH603 pKa = 6.73SYY605 pKa = 10.79TNVVNPSDD613 pKa = 3.51LTGVVGSLAGSGVVQSAAGNDD634 pKa = 4.09VINGGAGKK642 pKa = 10.04DD643 pKa = 3.38IIYY646 pKa = 10.23GDD648 pKa = 3.77VMNTDD653 pKa = 3.76VLRR656 pKa = 11.84AAAGLSAATYY666 pKa = 8.45PAGSGWGVFAALEE679 pKa = 4.25GTTTAFTTANDD690 pKa = 3.96LAGDD694 pKa = 4.66GPQWTRR700 pKa = 11.84ADD702 pKa = 3.91TIAYY706 pKa = 8.43IAAHH710 pKa = 5.41QSEE713 pKa = 4.54LSKK716 pKa = 11.12EE717 pKa = 4.19SGRR720 pKa = 11.84TGGDD724 pKa = 3.23DD725 pKa = 3.89TITGGMGDD733 pKa = 5.02DD734 pKa = 4.52IIFAQEE740 pKa = 4.06GNDD743 pKa = 3.59KK744 pKa = 10.39IYY746 pKa = 10.67YY747 pKa = 9.13AQGDD751 pKa = 3.92GKK753 pKa = 9.96DD754 pKa = 3.7TVDD757 pKa = 4.03GGSGSDD763 pKa = 3.14TLYY766 pKa = 10.66ISGATGTVTIAAATVGPDD784 pKa = 3.07IVPATGTNYY793 pKa = 10.23TDD795 pKa = 4.48IIVTMSDD802 pKa = 2.92GGSIRR807 pKa = 11.84MDD809 pKa = 3.26SVEE812 pKa = 5.58DD813 pKa = 3.47IDD815 pKa = 3.86ITAGPGGVNIQYY827 pKa = 10.08IGSLASTALDD837 pKa = 3.46TSTVTVHH844 pKa = 6.73GGTGDD849 pKa = 3.83DD850 pKa = 4.92VLDD853 pKa = 3.54LTQRR857 pKa = 11.84GNTDD861 pKa = 3.12PHH863 pKa = 8.1KK864 pKa = 10.99VIADD868 pKa = 4.06GGAQTTADD876 pKa = 3.18IVKK879 pKa = 10.29LDD881 pKa = 3.81FSVSEE886 pKa = 3.88ITAIQAISGGVKK898 pKa = 8.25ITHH901 pKa = 6.87AGGAGTITDD910 pKa = 3.61EE911 pKa = 4.31FTNFEE916 pKa = 4.15SFQFEE921 pKa = 4.25GGVTKK926 pKa = 10.07TLAEE930 pKa = 4.21VQNLDD935 pKa = 5.19LIPPTLTSITMNDD948 pKa = 3.14TALKK952 pKa = 10.28VGEE955 pKa = 4.25TSLVTVVFSEE965 pKa = 4.24KK966 pKa = 10.68VNNLEE971 pKa = 4.16VTDD974 pKa = 4.6FTAPGGALSNLSSADD989 pKa = 3.56GGQTWTMTFIPTTNSTSSGNQISLANNSYY1018 pKa = 10.58TDD1020 pKa = 3.14IAGNTGSGGTSAATYY1035 pKa = 10.53SIDD1038 pKa = 3.42TKK1040 pKa = 11.35APTVTINGDD1049 pKa = 3.25VDD1051 pKa = 3.91PVTGQQVVFDD1061 pKa = 4.11VVFSEE1066 pKa = 4.57SVTGFTGADD1075 pKa = 2.82ISFTGSTAPGSLSASVTGSGTTYY1098 pKa = 10.3QVTVTGMTGSGSVVASVIAGAALDD1122 pKa = 3.89AAGNASAASTSTDD1135 pKa = 2.97NAVAFTLNLPSFIIAPSDD1153 pKa = 3.51LQYY1156 pKa = 11.56DD1157 pKa = 4.01PTSSSDD1163 pKa = 3.09VTAFNRR1169 pKa = 11.84IMFSDD1174 pKa = 4.54PDD1176 pKa = 3.81TAGSVTVHH1184 pKa = 6.15LTRR1187 pKa = 11.84TSGDD1191 pKa = 3.31GTLSATSTSDD1201 pKa = 3.37VVVSGSSSDD1210 pKa = 3.86MILTGTVDD1218 pKa = 4.93AINAYY1223 pKa = 10.12LAGNKK1228 pKa = 7.22VTLDD1232 pKa = 3.47TQGNDD1237 pKa = 2.78TDD1239 pKa = 4.09VFTISIDD1246 pKa = 3.73DD1247 pKa = 3.79GTTTISQTNYY1257 pKa = 8.56TVSDD1261 pKa = 3.63VSFTSSGSANTNNYY1275 pKa = 10.23AGVNVYY1281 pKa = 7.53EE1282 pKa = 4.29TSLDD1286 pKa = 3.66SGNGNDD1292 pKa = 3.83TLYY1295 pKa = 10.72TSWSHH1300 pKa = 6.38LGSSATAYY1308 pKa = 10.5AGGGGTGDD1316 pKa = 3.97TINLIFTADD1325 pKa = 3.45QLQEE1329 pKa = 4.05ILSSGTTRR1337 pKa = 11.84DD1338 pKa = 3.54DD1339 pKa = 3.62LQAFLAAPTGDD1350 pKa = 3.64TLSLGSTSWRR1360 pKa = 11.84ASASGFEE1367 pKa = 4.16TANIQLADD1375 pKa = 4.46LYY1377 pKa = 10.39ATRR1380 pKa = 11.84DD1381 pKa = 3.24TTTPSNNYY1389 pKa = 9.71FDD1391 pKa = 6.13INTTWKK1397 pKa = 10.65SVITSSEE1404 pKa = 3.91LLGTNGDD1411 pKa = 3.8GNNNFIVASAAGQSLSGNGGNDD1433 pKa = 2.91VMVALNGGNTLNGGAGLDD1451 pKa = 3.86LLLGGSGSDD1460 pKa = 3.37TLIGGDD1466 pKa = 4.04GIDD1469 pKa = 3.7RR1470 pKa = 11.84LSGGGGIDD1478 pKa = 2.92HH1479 pKa = 6.96FRR1481 pKa = 11.84FNATSEE1487 pKa = 4.23AGDD1490 pKa = 4.16HH1491 pKa = 6.91IMDD1494 pKa = 4.92FVHH1497 pKa = 6.52GTDD1500 pKa = 4.56TIDD1503 pKa = 3.7LLAAAFGGGSGAIVSADD1520 pKa = 4.44LIQITDD1526 pKa = 3.63AQNPSTIDD1534 pKa = 2.94MGSAHH1539 pKa = 7.15FAYY1542 pKa = 10.04QQSTGSLYY1550 pKa = 10.59YY1551 pKa = 10.61DD1552 pKa = 4.05PNGGAADD1559 pKa = 3.73ANRR1562 pKa = 11.84ILLAVLDD1569 pKa = 3.79NHH1571 pKa = 7.05AALTATDD1578 pKa = 3.41VHH1580 pKa = 6.87KK1581 pKa = 11.27VV1582 pKa = 3.16
MM1 pKa = 7.45SATAAAGVLSNDD13 pKa = 3.08TFGADD18 pKa = 3.3GASVVGVAKK27 pKa = 9.44GTTNANLDD35 pKa = 3.55NTGTLGAGLHH45 pKa = 5.32GTYY48 pKa = 8.88GTLTLNADD56 pKa = 3.45GSYY59 pKa = 10.82SYY61 pKa = 11.01IRR63 pKa = 11.84DD64 pKa = 3.61ANSAGGVTDD73 pKa = 3.91TFTYY77 pKa = 9.47TIKK80 pKa = 11.15DD81 pKa = 3.23GDD83 pKa = 4.1GDD85 pKa = 4.18LSHH88 pKa = 6.71TTLTITLPDD97 pKa = 3.59ATPSYY102 pKa = 9.62TLPTAGNGVTTVYY115 pKa = 10.43EE116 pKa = 3.86AGLAVRR122 pKa = 11.84SGEE125 pKa = 4.0PAGSHH130 pKa = 6.16IGATNITATGVIGVSSVDD148 pKa = 3.52GVGSVSLNGQTVTGTSAGTATTVLSNSTGTLTAWYY183 pKa = 7.85EE184 pKa = 3.87AATSQIHH191 pKa = 4.33YY192 pKa = 9.06TYY194 pKa = 10.71TLLDD198 pKa = 3.46NTLNTSGTSISVPVGVTDD216 pKa = 5.78LDD218 pKa = 3.99GDD220 pKa = 4.22TPASGGNLVITIVDD234 pKa = 3.92DD235 pKa = 4.4APVAVSDD242 pKa = 4.05TNSLGTASVGSQASGNLIAGTITSGSGTGGADD274 pKa = 3.23TQGADD279 pKa = 3.41GAHH282 pKa = 6.65IGSVTGGASDD292 pKa = 3.64GSGGFNVAGTYY303 pKa = 7.48GTLHH307 pKa = 6.21VDD309 pKa = 3.26GNGAYY314 pKa = 10.22SYY316 pKa = 10.46TRR318 pKa = 11.84TSAAGGTDD326 pKa = 3.23TFHH329 pKa = 6.37YY330 pKa = 9.68TLTDD334 pKa = 3.38GDD336 pKa = 4.4GDD338 pKa = 3.58ISNQADD344 pKa = 3.77LTITLDD350 pKa = 3.24DD351 pKa = 3.65TGRR354 pKa = 11.84LFVGSNDD361 pKa = 4.2SDD363 pKa = 4.96DD364 pKa = 4.47NPNDD368 pKa = 3.47PAHH371 pKa = 6.22FVDD374 pKa = 4.2VSSNVTGVINGGGGNDD390 pKa = 4.39TIAGDD395 pKa = 3.96PGATPTISAGSSANIVLVLDD415 pKa = 4.2TTASLTAQQMSAMQQAVGALLNSLYY440 pKa = 10.96NSGAANVRR448 pKa = 11.84VSMYY452 pKa = 10.83AFGGGYY458 pKa = 7.04TQLGTYY464 pKa = 10.27NIVANSHH471 pKa = 6.01SNAAADD477 pKa = 3.62AAALVSALDD486 pKa = 3.65AANADD491 pKa = 3.71GSITNSGSRR500 pKa = 11.84LYY502 pKa = 10.38IGADD506 pKa = 3.15PGSNTSYY513 pKa = 11.62ADD515 pKa = 3.47GLDD518 pKa = 3.4AVIDD522 pKa = 3.92YY523 pKa = 7.37LTPGTSGGDD532 pKa = 3.43PVSAVPLTGASVNKK546 pKa = 10.15VIFISDD552 pKa = 4.2GEE554 pKa = 4.36PTTNATQTQINTITGTGAGQYY575 pKa = 10.1GYY577 pKa = 9.0TIEE580 pKa = 4.87AVGLNVAGSPNAVAALNALDD600 pKa = 4.64SGHH603 pKa = 6.73SYY605 pKa = 10.79TNVVNPSDD613 pKa = 3.51LTGVVGSLAGSGVVQSAAGNDD634 pKa = 4.09VINGGAGKK642 pKa = 10.04DD643 pKa = 3.38IIYY646 pKa = 10.23GDD648 pKa = 3.77VMNTDD653 pKa = 3.76VLRR656 pKa = 11.84AAAGLSAATYY666 pKa = 8.45PAGSGWGVFAALEE679 pKa = 4.25GTTTAFTTANDD690 pKa = 3.96LAGDD694 pKa = 4.66GPQWTRR700 pKa = 11.84ADD702 pKa = 3.91TIAYY706 pKa = 8.43IAAHH710 pKa = 5.41QSEE713 pKa = 4.54LSKK716 pKa = 11.12EE717 pKa = 4.19SGRR720 pKa = 11.84TGGDD724 pKa = 3.23DD725 pKa = 3.89TITGGMGDD733 pKa = 5.02DD734 pKa = 4.52IIFAQEE740 pKa = 4.06GNDD743 pKa = 3.59KK744 pKa = 10.39IYY746 pKa = 10.67YY747 pKa = 9.13AQGDD751 pKa = 3.92GKK753 pKa = 9.96DD754 pKa = 3.7TVDD757 pKa = 4.03GGSGSDD763 pKa = 3.14TLYY766 pKa = 10.66ISGATGTVTIAAATVGPDD784 pKa = 3.07IVPATGTNYY793 pKa = 10.23TDD795 pKa = 4.48IIVTMSDD802 pKa = 2.92GGSIRR807 pKa = 11.84MDD809 pKa = 3.26SVEE812 pKa = 5.58DD813 pKa = 3.47IDD815 pKa = 3.86ITAGPGGVNIQYY827 pKa = 10.08IGSLASTALDD837 pKa = 3.46TSTVTVHH844 pKa = 6.73GGTGDD849 pKa = 3.83DD850 pKa = 4.92VLDD853 pKa = 3.54LTQRR857 pKa = 11.84GNTDD861 pKa = 3.12PHH863 pKa = 8.1KK864 pKa = 10.99VIADD868 pKa = 4.06GGAQTTADD876 pKa = 3.18IVKK879 pKa = 10.29LDD881 pKa = 3.81FSVSEE886 pKa = 3.88ITAIQAISGGVKK898 pKa = 8.25ITHH901 pKa = 6.87AGGAGTITDD910 pKa = 3.61EE911 pKa = 4.31FTNFEE916 pKa = 4.15SFQFEE921 pKa = 4.25GGVTKK926 pKa = 10.07TLAEE930 pKa = 4.21VQNLDD935 pKa = 5.19LIPPTLTSITMNDD948 pKa = 3.14TALKK952 pKa = 10.28VGEE955 pKa = 4.25TSLVTVVFSEE965 pKa = 4.24KK966 pKa = 10.68VNNLEE971 pKa = 4.16VTDD974 pKa = 4.6FTAPGGALSNLSSADD989 pKa = 3.56GGQTWTMTFIPTTNSTSSGNQISLANNSYY1018 pKa = 10.58TDD1020 pKa = 3.14IAGNTGSGGTSAATYY1035 pKa = 10.53SIDD1038 pKa = 3.42TKK1040 pKa = 11.35APTVTINGDD1049 pKa = 3.25VDD1051 pKa = 3.91PVTGQQVVFDD1061 pKa = 4.11VVFSEE1066 pKa = 4.57SVTGFTGADD1075 pKa = 2.82ISFTGSTAPGSLSASVTGSGTTYY1098 pKa = 10.3QVTVTGMTGSGSVVASVIAGAALDD1122 pKa = 3.89AAGNASAASTSTDD1135 pKa = 2.97NAVAFTLNLPSFIIAPSDD1153 pKa = 3.51LQYY1156 pKa = 11.56DD1157 pKa = 4.01PTSSSDD1163 pKa = 3.09VTAFNRR1169 pKa = 11.84IMFSDD1174 pKa = 4.54PDD1176 pKa = 3.81TAGSVTVHH1184 pKa = 6.15LTRR1187 pKa = 11.84TSGDD1191 pKa = 3.31GTLSATSTSDD1201 pKa = 3.37VVVSGSSSDD1210 pKa = 3.86MILTGTVDD1218 pKa = 4.93AINAYY1223 pKa = 10.12LAGNKK1228 pKa = 7.22VTLDD1232 pKa = 3.47TQGNDD1237 pKa = 2.78TDD1239 pKa = 4.09VFTISIDD1246 pKa = 3.73DD1247 pKa = 3.79GTTTISQTNYY1257 pKa = 8.56TVSDD1261 pKa = 3.63VSFTSSGSANTNNYY1275 pKa = 10.23AGVNVYY1281 pKa = 7.53EE1282 pKa = 4.29TSLDD1286 pKa = 3.66SGNGNDD1292 pKa = 3.83TLYY1295 pKa = 10.72TSWSHH1300 pKa = 6.38LGSSATAYY1308 pKa = 10.5AGGGGTGDD1316 pKa = 3.97TINLIFTADD1325 pKa = 3.45QLQEE1329 pKa = 4.05ILSSGTTRR1337 pKa = 11.84DD1338 pKa = 3.54DD1339 pKa = 3.62LQAFLAAPTGDD1350 pKa = 3.64TLSLGSTSWRR1360 pKa = 11.84ASASGFEE1367 pKa = 4.16TANIQLADD1375 pKa = 4.46LYY1377 pKa = 10.39ATRR1380 pKa = 11.84DD1381 pKa = 3.24TTTPSNNYY1389 pKa = 9.71FDD1391 pKa = 6.13INTTWKK1397 pKa = 10.65SVITSSEE1404 pKa = 3.91LLGTNGDD1411 pKa = 3.8GNNNFIVASAAGQSLSGNGGNDD1433 pKa = 2.91VMVALNGGNTLNGGAGLDD1451 pKa = 3.86LLLGGSGSDD1460 pKa = 3.37TLIGGDD1466 pKa = 4.04GIDD1469 pKa = 3.7RR1470 pKa = 11.84LSGGGGIDD1478 pKa = 2.92HH1479 pKa = 6.96FRR1481 pKa = 11.84FNATSEE1487 pKa = 4.23AGDD1490 pKa = 4.16HH1491 pKa = 6.91IMDD1494 pKa = 4.92FVHH1497 pKa = 6.52GTDD1500 pKa = 4.56TIDD1503 pKa = 3.7LLAAAFGGGSGAIVSADD1520 pKa = 4.44LIQITDD1526 pKa = 3.63AQNPSTIDD1534 pKa = 2.94MGSAHH1539 pKa = 7.15FAYY1542 pKa = 10.04QQSTGSLYY1550 pKa = 10.59YY1551 pKa = 10.61DD1552 pKa = 4.05PNGGAADD1559 pKa = 3.73ANRR1562 pKa = 11.84ILLAVLDD1569 pKa = 3.79NHH1571 pKa = 7.05AALTATDD1578 pKa = 3.41VHH1580 pKa = 6.87KK1581 pKa = 11.27VV1582 pKa = 3.16
Molecular weight: 157.52 kDa
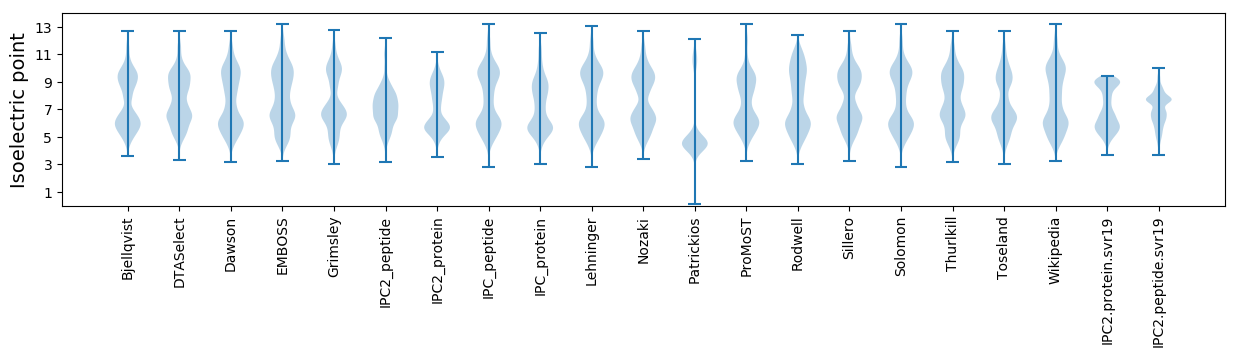
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F7QFR9|F7QFR9_9BRAD Uncharacterized protein OS=Bradyrhizobiaceae bacterium SG-6C OX=709797 GN=CSIRO_0292 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84LATAGGRR28 pKa = 11.84KK29 pKa = 8.95VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84LATAGGRR28 pKa = 11.84KK29 pKa = 8.95VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1249219 |
37 |
2736 |
294.9 |
31.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.254 ± 0.051 | 0.815 ± 0.012 |
5.598 ± 0.031 | 5.192 ± 0.031 |
3.836 ± 0.023 | 8.397 ± 0.052 |
1.961 ± 0.02 | 5.589 ± 0.029 |
4.125 ± 0.031 | 9.592 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.591 ± 0.018 | 2.906 ± 0.023 |
5.193 ± 0.03 | 3.131 ± 0.022 |
6.662 ± 0.034 | 5.676 ± 0.029 |
5.485 ± 0.03 | 7.465 ± 0.033 |
1.255 ± 0.014 | 2.278 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
