
Asticcacaulis taihuensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Asticcacaulis
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
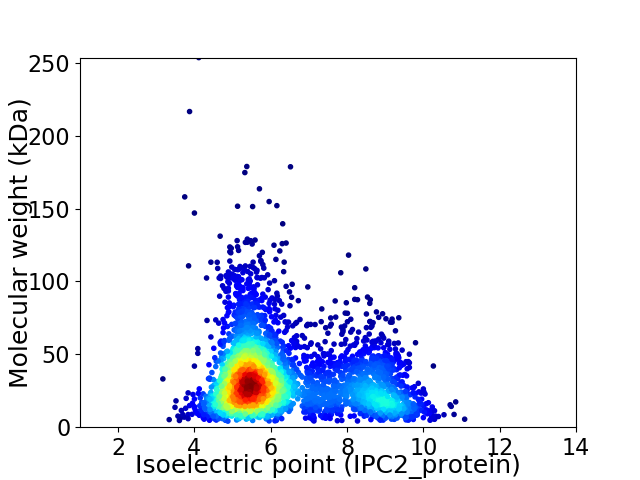
Virtual 2D-PAGE plot for 3569 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G4PFH0|A0A1G4PFH0_9CAUL Probable cytosol aminopeptidase OS=Asticcacaulis taihuensis OX=260084 GN=pepA PE=3 SV=1
MM1 pKa = 7.94RR2 pKa = 11.84KK3 pKa = 9.49HH4 pKa = 6.39SLLTATALGLVLGSGGLGTQALAEE28 pKa = 4.3TSISTATTTPLVTSSAGDD46 pKa = 3.47LTVTSDD52 pKa = 3.65GSITLTSGTAITVDD66 pKa = 3.76SDD68 pKa = 3.32NSLDD72 pKa = 3.92FEE74 pKa = 4.8GSIAMSGSEE83 pKa = 4.08SSSTGILITDD93 pKa = 4.52FPDD96 pKa = 3.3RR97 pKa = 11.84TKK99 pKa = 11.42GLVLTGDD106 pKa = 3.21ITVTDD111 pKa = 5.36DD112 pKa = 3.31YY113 pKa = 10.57TASDD117 pKa = 3.8TTSLDD122 pKa = 3.38GTEE125 pKa = 5.26DD126 pKa = 4.43GYY128 pKa = 9.82TDD130 pKa = 4.11APWAEE135 pKa = 3.93GTGRR139 pKa = 11.84YY140 pKa = 9.12GIHH143 pKa = 5.83STGASPFVGDD153 pKa = 4.08VYY155 pKa = 10.28ITSDD159 pKa = 3.38SVIDD163 pKa = 3.71VEE165 pKa = 5.47GNQSYY170 pKa = 10.57GIRR173 pKa = 11.84FEE175 pKa = 4.15NQIKK179 pKa = 10.32GAFTYY184 pKa = 10.5DD185 pKa = 2.98GAMTLIGDD193 pKa = 3.81NSTGISLEE201 pKa = 3.9KK202 pKa = 10.59GVTGNVYY209 pKa = 10.49LSGSVGVLGANASAITLAGDD229 pKa = 3.93LGGNLIIDD237 pKa = 4.26GSYY240 pKa = 10.77SGTAYY245 pKa = 10.8SSTSALTQAVYY256 pKa = 11.06EE257 pKa = 4.1NLIPANNLLQVGPLVSIAGNVANGVLFGASVTSTDD292 pKa = 5.16DD293 pKa = 3.6SNTDD297 pKa = 3.12EE298 pKa = 5.92DD299 pKa = 5.48GDD301 pKa = 4.21GLTDD305 pKa = 4.04TVQSTASLTQYY316 pKa = 10.97GSAPALAIGSASSDD330 pKa = 2.86ITLGGLTYY338 pKa = 10.92ASTAIDD344 pKa = 4.22PPSVNYY350 pKa = 10.56GLLNRR355 pKa = 11.84GSIGAYY361 pKa = 9.15GVHH364 pKa = 6.82PGVNATALQIGGTGYY379 pKa = 7.61PTTIEE384 pKa = 4.03NGIGNAGSITASSYY398 pKa = 11.06GGNATAISLLSGATTPRR415 pKa = 11.84LDD417 pKa = 3.3IDD419 pKa = 3.89GTVTATTTRR428 pKa = 11.84YY429 pKa = 7.8LTSAEE434 pKa = 4.15DD435 pKa = 3.74ADD437 pKa = 4.28GNTVYY442 pKa = 10.14TVSNVGEE449 pKa = 4.05ATAVSIASGATLPAINVAASSGIYY473 pKa = 10.01ASSSGSTGSATAISDD488 pKa = 3.51TSGTLTSISNSGTISATITASDD510 pKa = 4.3DD511 pKa = 4.42DD512 pKa = 5.21GDD514 pKa = 4.18GTTDD518 pKa = 4.07TITGKK523 pKa = 8.83ATAIDD528 pKa = 3.84VSTNTTGVTITQTDD542 pKa = 3.51NNATDD547 pKa = 3.62SDD549 pKa = 4.22DD550 pKa = 5.0DD551 pKa = 3.91EE552 pKa = 7.45AIAAPYY558 pKa = 9.94IYY560 pKa = 10.88GNILFGSGNDD570 pKa = 3.73TLSSSGGYY578 pKa = 9.63IYY580 pKa = 11.2GNVDD584 pKa = 3.06YY585 pKa = 10.87GAGTGSFSLTNDD597 pKa = 2.76AVYY600 pKa = 10.42LGKK603 pKa = 8.94LTSSGDD609 pKa = 3.0IAMDD613 pKa = 3.45IDD615 pKa = 4.05SGASAGLLAGSSVRR629 pKa = 11.84MSTLHH634 pKa = 6.02VGSDD638 pKa = 3.07SSMALTLAVDD648 pKa = 4.27TPTVPILSGSGAVAFDD664 pKa = 4.83DD665 pKa = 4.72GAKK668 pKa = 10.42LYY670 pKa = 8.65LTLDD674 pKa = 4.56KK675 pKa = 10.78ILSTPTSFTVLTGSSISLGDD695 pKa = 3.6MTTSTLDD702 pKa = 3.22GYY704 pKa = 10.55IPYY707 pKa = 9.88LYY709 pKa = 10.69HH710 pKa = 7.96SDD712 pKa = 3.35LTLNDD717 pKa = 3.51TDD719 pKa = 4.16TVLTANFRR727 pKa = 11.84LKK729 pKa = 9.33TQGEE733 pKa = 4.2AGYY736 pKa = 10.88SSNQYY741 pKa = 9.52AALMPVLVVVAEE753 pKa = 4.27DD754 pKa = 3.13SGARR758 pKa = 11.84SSLLAATDD766 pKa = 3.52KK767 pKa = 11.41DD768 pKa = 4.61AFDD771 pKa = 3.55QVYY774 pKa = 9.77NQYY777 pKa = 11.0LPDD780 pKa = 3.63YY781 pKa = 9.84SGEE784 pKa = 4.05NLISLSVGSASLNRR798 pKa = 11.84SLSNLTLIPDD808 pKa = 4.06NNGGQYY814 pKa = 9.34WLQEE818 pKa = 3.52YY819 pKa = 10.18GYY821 pKa = 9.9AINRR825 pKa = 11.84AYY827 pKa = 11.08SDD829 pKa = 3.34TAGFKK834 pKa = 9.49STGFSFAGGRR844 pKa = 11.84EE845 pKa = 3.84KK846 pKa = 10.4QVYY849 pKa = 7.91GNQMVGTYY857 pKa = 10.56LSITSASPRR866 pKa = 11.84DD867 pKa = 4.0TFALAAEE874 pKa = 4.72GMSNSDD880 pKa = 3.69LTIGAYY886 pKa = 8.5WRR888 pKa = 11.84LNTSGLKK895 pKa = 9.94AWLHH899 pKa = 6.25AGAGYY904 pKa = 8.18DD905 pKa = 3.55TFKK908 pKa = 10.05STRR911 pKa = 11.84NILTTTVDD919 pKa = 3.52HH920 pKa = 6.32VATAKK925 pKa = 9.47WDD927 pKa = 4.13GYY929 pKa = 9.98SVSAGTGASYY939 pKa = 11.52DD940 pKa = 3.65FLLGKK945 pKa = 9.13WAVTPEE951 pKa = 3.95VLADD955 pKa = 3.39YY956 pKa = 10.28YY957 pKa = 10.9QLHH960 pKa = 6.67EE961 pKa = 4.62NDD963 pKa = 3.6HH964 pKa = 6.07TEE966 pKa = 4.13SGGGDD971 pKa = 3.66YY972 pKa = 11.07FDD974 pKa = 4.31LTVGARR980 pKa = 11.84DD981 pKa = 3.52GHH983 pKa = 6.41LLSSTALLNLSYY995 pKa = 9.62RR996 pKa = 11.84TSFLKK1001 pKa = 10.51PEE1003 pKa = 3.68MWVGYY1008 pKa = 8.66KK1009 pKa = 10.19QNISATLPDD1018 pKa = 3.66TVANFTGGDD1027 pKa = 3.46SFTLVGGNIEE1037 pKa = 4.2GGGPVAGFRR1046 pKa = 11.84LSADD1050 pKa = 3.05NPYY1053 pKa = 10.9SYY1055 pKa = 11.11FSIEE1059 pKa = 3.92AEE1061 pKa = 4.07YY1062 pKa = 11.03QEE1064 pKa = 4.58LPEE1067 pKa = 4.07YY1068 pKa = 10.46TNTSVSLRR1076 pKa = 11.84TRR1078 pKa = 11.84FQFF1081 pKa = 4.0
MM1 pKa = 7.94RR2 pKa = 11.84KK3 pKa = 9.49HH4 pKa = 6.39SLLTATALGLVLGSGGLGTQALAEE28 pKa = 4.3TSISTATTTPLVTSSAGDD46 pKa = 3.47LTVTSDD52 pKa = 3.65GSITLTSGTAITVDD66 pKa = 3.76SDD68 pKa = 3.32NSLDD72 pKa = 3.92FEE74 pKa = 4.8GSIAMSGSEE83 pKa = 4.08SSSTGILITDD93 pKa = 4.52FPDD96 pKa = 3.3RR97 pKa = 11.84TKK99 pKa = 11.42GLVLTGDD106 pKa = 3.21ITVTDD111 pKa = 5.36DD112 pKa = 3.31YY113 pKa = 10.57TASDD117 pKa = 3.8TTSLDD122 pKa = 3.38GTEE125 pKa = 5.26DD126 pKa = 4.43GYY128 pKa = 9.82TDD130 pKa = 4.11APWAEE135 pKa = 3.93GTGRR139 pKa = 11.84YY140 pKa = 9.12GIHH143 pKa = 5.83STGASPFVGDD153 pKa = 4.08VYY155 pKa = 10.28ITSDD159 pKa = 3.38SVIDD163 pKa = 3.71VEE165 pKa = 5.47GNQSYY170 pKa = 10.57GIRR173 pKa = 11.84FEE175 pKa = 4.15NQIKK179 pKa = 10.32GAFTYY184 pKa = 10.5DD185 pKa = 2.98GAMTLIGDD193 pKa = 3.81NSTGISLEE201 pKa = 3.9KK202 pKa = 10.59GVTGNVYY209 pKa = 10.49LSGSVGVLGANASAITLAGDD229 pKa = 3.93LGGNLIIDD237 pKa = 4.26GSYY240 pKa = 10.77SGTAYY245 pKa = 10.8SSTSALTQAVYY256 pKa = 11.06EE257 pKa = 4.1NLIPANNLLQVGPLVSIAGNVANGVLFGASVTSTDD292 pKa = 5.16DD293 pKa = 3.6SNTDD297 pKa = 3.12EE298 pKa = 5.92DD299 pKa = 5.48GDD301 pKa = 4.21GLTDD305 pKa = 4.04TVQSTASLTQYY316 pKa = 10.97GSAPALAIGSASSDD330 pKa = 2.86ITLGGLTYY338 pKa = 10.92ASTAIDD344 pKa = 4.22PPSVNYY350 pKa = 10.56GLLNRR355 pKa = 11.84GSIGAYY361 pKa = 9.15GVHH364 pKa = 6.82PGVNATALQIGGTGYY379 pKa = 7.61PTTIEE384 pKa = 4.03NGIGNAGSITASSYY398 pKa = 11.06GGNATAISLLSGATTPRR415 pKa = 11.84LDD417 pKa = 3.3IDD419 pKa = 3.89GTVTATTTRR428 pKa = 11.84YY429 pKa = 7.8LTSAEE434 pKa = 4.15DD435 pKa = 3.74ADD437 pKa = 4.28GNTVYY442 pKa = 10.14TVSNVGEE449 pKa = 4.05ATAVSIASGATLPAINVAASSGIYY473 pKa = 10.01ASSSGSTGSATAISDD488 pKa = 3.51TSGTLTSISNSGTISATITASDD510 pKa = 4.3DD511 pKa = 4.42DD512 pKa = 5.21GDD514 pKa = 4.18GTTDD518 pKa = 4.07TITGKK523 pKa = 8.83ATAIDD528 pKa = 3.84VSTNTTGVTITQTDD542 pKa = 3.51NNATDD547 pKa = 3.62SDD549 pKa = 4.22DD550 pKa = 5.0DD551 pKa = 3.91EE552 pKa = 7.45AIAAPYY558 pKa = 9.94IYY560 pKa = 10.88GNILFGSGNDD570 pKa = 3.73TLSSSGGYY578 pKa = 9.63IYY580 pKa = 11.2GNVDD584 pKa = 3.06YY585 pKa = 10.87GAGTGSFSLTNDD597 pKa = 2.76AVYY600 pKa = 10.42LGKK603 pKa = 8.94LTSSGDD609 pKa = 3.0IAMDD613 pKa = 3.45IDD615 pKa = 4.05SGASAGLLAGSSVRR629 pKa = 11.84MSTLHH634 pKa = 6.02VGSDD638 pKa = 3.07SSMALTLAVDD648 pKa = 4.27TPTVPILSGSGAVAFDD664 pKa = 4.83DD665 pKa = 4.72GAKK668 pKa = 10.42LYY670 pKa = 8.65LTLDD674 pKa = 4.56KK675 pKa = 10.78ILSTPTSFTVLTGSSISLGDD695 pKa = 3.6MTTSTLDD702 pKa = 3.22GYY704 pKa = 10.55IPYY707 pKa = 9.88LYY709 pKa = 10.69HH710 pKa = 7.96SDD712 pKa = 3.35LTLNDD717 pKa = 3.51TDD719 pKa = 4.16TVLTANFRR727 pKa = 11.84LKK729 pKa = 9.33TQGEE733 pKa = 4.2AGYY736 pKa = 10.88SSNQYY741 pKa = 9.52AALMPVLVVVAEE753 pKa = 4.27DD754 pKa = 3.13SGARR758 pKa = 11.84SSLLAATDD766 pKa = 3.52KK767 pKa = 11.41DD768 pKa = 4.61AFDD771 pKa = 3.55QVYY774 pKa = 9.77NQYY777 pKa = 11.0LPDD780 pKa = 3.63YY781 pKa = 9.84SGEE784 pKa = 4.05NLISLSVGSASLNRR798 pKa = 11.84SLSNLTLIPDD808 pKa = 4.06NNGGQYY814 pKa = 9.34WLQEE818 pKa = 3.52YY819 pKa = 10.18GYY821 pKa = 9.9AINRR825 pKa = 11.84AYY827 pKa = 11.08SDD829 pKa = 3.34TAGFKK834 pKa = 9.49STGFSFAGGRR844 pKa = 11.84EE845 pKa = 3.84KK846 pKa = 10.4QVYY849 pKa = 7.91GNQMVGTYY857 pKa = 10.56LSITSASPRR866 pKa = 11.84DD867 pKa = 4.0TFALAAEE874 pKa = 4.72GMSNSDD880 pKa = 3.69LTIGAYY886 pKa = 8.5WRR888 pKa = 11.84LNTSGLKK895 pKa = 9.94AWLHH899 pKa = 6.25AGAGYY904 pKa = 8.18DD905 pKa = 3.55TFKK908 pKa = 10.05STRR911 pKa = 11.84NILTTTVDD919 pKa = 3.52HH920 pKa = 6.32VATAKK925 pKa = 9.47WDD927 pKa = 4.13GYY929 pKa = 9.98SVSAGTGASYY939 pKa = 11.52DD940 pKa = 3.65FLLGKK945 pKa = 9.13WAVTPEE951 pKa = 3.95VLADD955 pKa = 3.39YY956 pKa = 10.28YY957 pKa = 10.9QLHH960 pKa = 6.67EE961 pKa = 4.62NDD963 pKa = 3.6HH964 pKa = 6.07TEE966 pKa = 4.13SGGGDD971 pKa = 3.66YY972 pKa = 11.07FDD974 pKa = 4.31LTVGARR980 pKa = 11.84DD981 pKa = 3.52GHH983 pKa = 6.41LLSSTALLNLSYY995 pKa = 9.62RR996 pKa = 11.84TSFLKK1001 pKa = 10.51PEE1003 pKa = 3.68MWVGYY1008 pKa = 8.66KK1009 pKa = 10.19QNISATLPDD1018 pKa = 3.66TVANFTGGDD1027 pKa = 3.46SFTLVGGNIEE1037 pKa = 4.2GGGPVAGFRR1046 pKa = 11.84LSADD1050 pKa = 3.05NPYY1053 pKa = 10.9SYY1055 pKa = 11.11FSIEE1059 pKa = 3.92AEE1061 pKa = 4.07YY1062 pKa = 11.03QEE1064 pKa = 4.58LPEE1067 pKa = 4.07YY1068 pKa = 10.46TNTSVSLRR1076 pKa = 11.84TRR1078 pKa = 11.84FQFF1081 pKa = 4.0
Molecular weight: 110.73 kDa
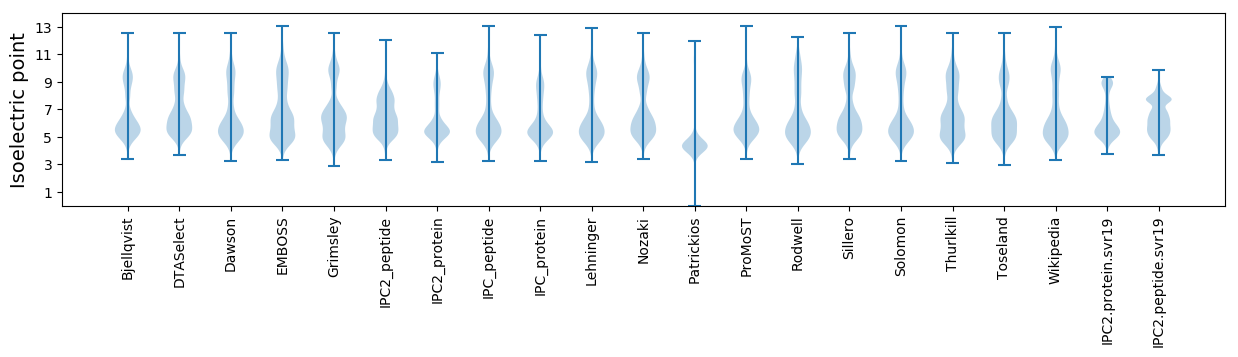
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G4PWI4|A0A1G4PWI4_9CAUL EF hand OS=Asticcacaulis taihuensis OX=260084 GN=SAMN02927928_0703 PE=4 SV=1
MM1 pKa = 7.29KK2 pKa = 10.41RR3 pKa = 11.84SLTNRR8 pKa = 11.84ATQVLDD14 pKa = 4.01LLPFAQLLTRR24 pKa = 11.84RR25 pKa = 11.84PPSGKK30 pKa = 10.17LLTAAALVNSPMANRR45 pKa = 11.84LPPRR49 pKa = 11.84LVGFAKK55 pKa = 10.26IAVAALAVGSVIAGARR71 pKa = 11.84QRR73 pKa = 11.84PGFKK77 pKa = 8.66WHH79 pKa = 5.5QRR81 pKa = 3.32
MM1 pKa = 7.29KK2 pKa = 10.41RR3 pKa = 11.84SLTNRR8 pKa = 11.84ATQVLDD14 pKa = 4.01LLPFAQLLTRR24 pKa = 11.84RR25 pKa = 11.84PPSGKK30 pKa = 10.17LLTAAALVNSPMANRR45 pKa = 11.84LPPRR49 pKa = 11.84LVGFAKK55 pKa = 10.26IAVAALAVGSVIAGARR71 pKa = 11.84QRR73 pKa = 11.84PGFKK77 pKa = 8.66WHH79 pKa = 5.5QRR81 pKa = 3.32
Molecular weight: 8.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1177575 |
39 |
2501 |
329.9 |
35.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.717 ± 0.05 | 0.812 ± 0.012 |
6.022 ± 0.032 | 5.244 ± 0.039 |
3.791 ± 0.026 | 8.23 ± 0.034 |
2.119 ± 0.019 | 5.319 ± 0.028 |
4.144 ± 0.028 | 9.934 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.516 ± 0.021 | 3.094 ± 0.029 |
5.062 ± 0.036 | 3.419 ± 0.025 |
5.971 ± 0.039 | 5.709 ± 0.035 |
5.772 ± 0.049 | 6.972 ± 0.032 |
1.435 ± 0.018 | 2.718 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
