
Tremella mesenterica (Jelly fungus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Tremellomycetes; Tremellales; Tremellaceae; Tremella
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
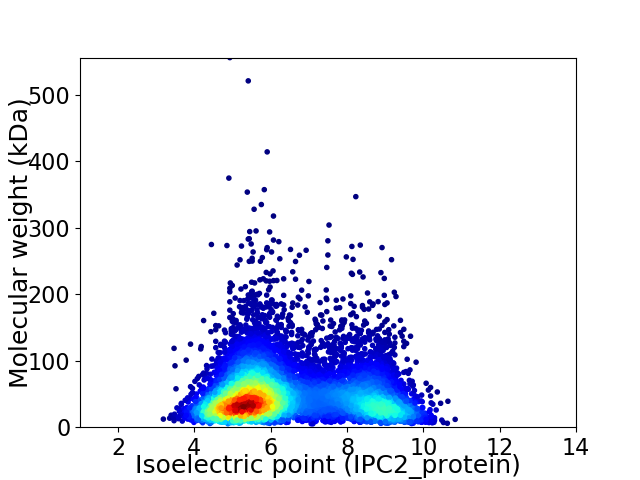
Virtual 2D-PAGE plot for 8061 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
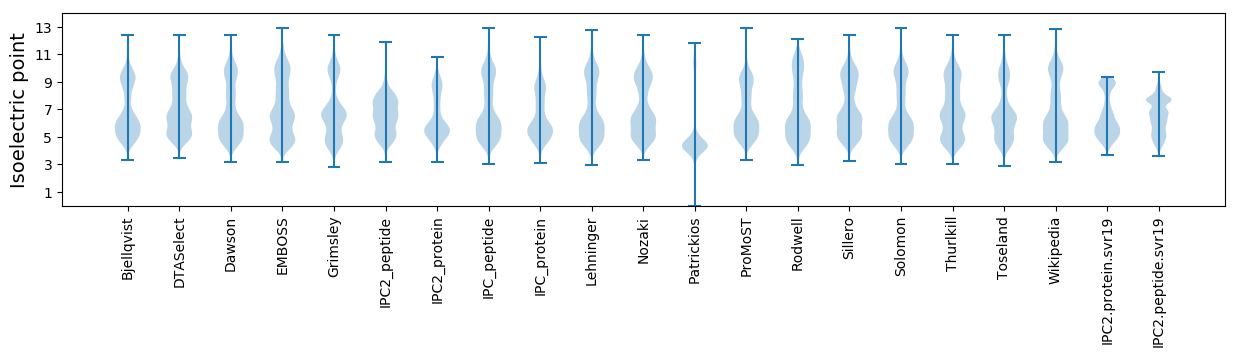
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q1BRF8|A0A4Q1BRF8_TREME Uncharacterized protein OS=Tremella mesenterica OX=5217 GN=M231_02218 PE=4 SV=1
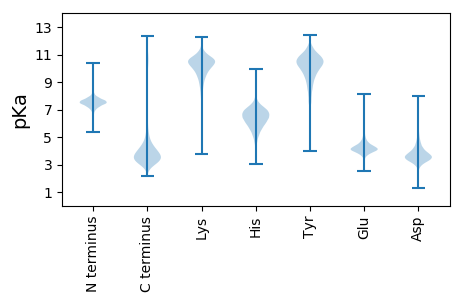
MM1 pKa = 8.28DD2 pKa = 6.76DD3 pKa = 5.06DD4 pKa = 3.95TTGWLRR10 pKa = 11.84YY11 pKa = 9.12SVQPLPEE18 pKa = 5.1DD19 pKa = 3.62SNNTEE24 pKa = 4.48GCPSAYY30 pKa = 10.3YY31 pKa = 10.1VIQPLADD38 pKa = 4.97DD39 pKa = 4.48GTHH42 pKa = 5.67LTSTVHH48 pKa = 7.11LKK50 pKa = 10.4QEE52 pKa = 4.63DD53 pKa = 3.79KK54 pKa = 11.33DD55 pKa = 4.0GTTMTTVKK63 pKa = 10.71GGAFTLNSFGPEE75 pKa = 4.06FEE77 pKa = 6.34AIDD80 pKa = 3.73PTSIRR85 pKa = 11.84DD86 pKa = 3.64GSGLGEE92 pKa = 4.59NILFQYY98 pKa = 10.09RR99 pKa = 11.84PSSFIDD105 pKa = 4.66FYY107 pKa = 11.71DD108 pKa = 4.18DD109 pKa = 3.52NNVIADD115 pKa = 4.51AVACLQDD122 pKa = 3.16IPMPSSFSLGVVDD135 pKa = 3.67WTVCSTPATTLGNQGQWLRR154 pKa = 11.84CTEE157 pKa = 4.0SMTVCAITPEE167 pKa = 4.13VGGGGEE173 pKa = 4.2RR174 pKa = 11.84GQEE177 pKa = 4.09SLVGAVEE184 pKa = 4.06VMEE187 pKa = 4.33II188 pKa = 3.78
MM1 pKa = 8.28DD2 pKa = 6.76DD3 pKa = 5.06DD4 pKa = 3.95TTGWLRR10 pKa = 11.84YY11 pKa = 9.12SVQPLPEE18 pKa = 5.1DD19 pKa = 3.62SNNTEE24 pKa = 4.48GCPSAYY30 pKa = 10.3YY31 pKa = 10.1VIQPLADD38 pKa = 4.97DD39 pKa = 4.48GTHH42 pKa = 5.67LTSTVHH48 pKa = 7.11LKK50 pKa = 10.4QEE52 pKa = 4.63DD53 pKa = 3.79KK54 pKa = 11.33DD55 pKa = 4.0GTTMTTVKK63 pKa = 10.71GGAFTLNSFGPEE75 pKa = 4.06FEE77 pKa = 6.34AIDD80 pKa = 3.73PTSIRR85 pKa = 11.84DD86 pKa = 3.64GSGLGEE92 pKa = 4.59NILFQYY98 pKa = 10.09RR99 pKa = 11.84PSSFIDD105 pKa = 4.66FYY107 pKa = 11.71DD108 pKa = 4.18DD109 pKa = 3.52NNVIADD115 pKa = 4.51AVACLQDD122 pKa = 3.16IPMPSSFSLGVVDD135 pKa = 3.67WTVCSTPATTLGNQGQWLRR154 pKa = 11.84CTEE157 pKa = 4.0SMTVCAITPEE167 pKa = 4.13VGGGGEE173 pKa = 4.2RR174 pKa = 11.84GQEE177 pKa = 4.09SLVGAVEE184 pKa = 4.06VMEE187 pKa = 4.33II188 pKa = 3.78
Molecular weight: 20.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q1BIT3|A0A4Q1BIT3_TREME Uncharacterized protein OS=Tremella mesenterica OX=5217 GN=M231_05171 PE=4 SV=1
MM1 pKa = 7.62CIVYY5 pKa = 9.6IRR7 pKa = 11.84KK8 pKa = 9.48VSTTSRR14 pKa = 11.84SLPVARR20 pKa = 11.84SPLPPAARR28 pKa = 11.84RR29 pKa = 11.84SVFATEE35 pKa = 4.54SPSPSDD41 pKa = 4.23EE42 pKa = 4.62LPGLPPPLPPSVSFRR57 pKa = 11.84AARR60 pKa = 11.84PGAPLWEE67 pKa = 4.95LSPSPPPRR75 pKa = 11.84GPRR78 pKa = 11.84PPRR81 pKa = 11.84GPRR84 pKa = 11.84PPRR87 pKa = 11.84APPVVNLCSPSPIVPGAFVVPAPPSPPRR115 pKa = 11.84PSPAPPLSSSSSSEE129 pKa = 3.72EE130 pKa = 3.85VPPRR134 pKa = 11.84TQEE137 pKa = 4.09EE138 pKa = 4.5IIADD142 pKa = 3.74LRR144 pKa = 11.84EE145 pKa = 4.16LVEE148 pKa = 5.53VLTLDD153 pKa = 3.43NLKK156 pKa = 9.32TEE158 pKa = 4.09KK159 pKa = 10.35RR160 pKa = 11.84LAAMTVVALRR170 pKa = 11.84AADD173 pKa = 3.56MMDD176 pKa = 3.22VQGVIIRR183 pKa = 11.84RR184 pKa = 11.84FLNQIRR190 pKa = 11.84LSTRR194 pKa = 11.84NLSKK198 pKa = 10.52KK199 pKa = 9.81SSHH202 pKa = 6.62IGSATHH208 pKa = 7.0SLLLQWNEE216 pKa = 3.39LLLRR220 pKa = 11.84TGCHH224 pKa = 6.78AIFGHH229 pKa = 6.0KK230 pKa = 9.61FWGEE234 pKa = 3.59INDD237 pKa = 4.36TIRR240 pKa = 11.84QKK242 pKa = 10.91EE243 pKa = 3.74RR244 pKa = 11.84DD245 pKa = 3.61LTGVAVAGPSTQGLRR260 pKa = 11.84DD261 pKa = 3.58EE262 pKa = 4.51VRR264 pKa = 11.84AASEE268 pKa = 4.45TFWDD272 pKa = 4.02ANWRR276 pKa = 11.84MRR278 pKa = 11.84NIRR281 pKa = 11.84GVDD284 pKa = 3.37VSLGQGTRR292 pKa = 11.84RR293 pKa = 11.84GTTANYY299 pKa = 9.7AVVISRR305 pKa = 11.84VCVEE309 pKa = 4.28LLGGSSPP316 pKa = 4.02
MM1 pKa = 7.62CIVYY5 pKa = 9.6IRR7 pKa = 11.84KK8 pKa = 9.48VSTTSRR14 pKa = 11.84SLPVARR20 pKa = 11.84SPLPPAARR28 pKa = 11.84RR29 pKa = 11.84SVFATEE35 pKa = 4.54SPSPSDD41 pKa = 4.23EE42 pKa = 4.62LPGLPPPLPPSVSFRR57 pKa = 11.84AARR60 pKa = 11.84PGAPLWEE67 pKa = 4.95LSPSPPPRR75 pKa = 11.84GPRR78 pKa = 11.84PPRR81 pKa = 11.84GPRR84 pKa = 11.84PPRR87 pKa = 11.84APPVVNLCSPSPIVPGAFVVPAPPSPPRR115 pKa = 11.84PSPAPPLSSSSSSEE129 pKa = 3.72EE130 pKa = 3.85VPPRR134 pKa = 11.84TQEE137 pKa = 4.09EE138 pKa = 4.5IIADD142 pKa = 3.74LRR144 pKa = 11.84EE145 pKa = 4.16LVEE148 pKa = 5.53VLTLDD153 pKa = 3.43NLKK156 pKa = 9.32TEE158 pKa = 4.09KK159 pKa = 10.35RR160 pKa = 11.84LAAMTVVALRR170 pKa = 11.84AADD173 pKa = 3.56MMDD176 pKa = 3.22VQGVIIRR183 pKa = 11.84RR184 pKa = 11.84FLNQIRR190 pKa = 11.84LSTRR194 pKa = 11.84NLSKK198 pKa = 10.52KK199 pKa = 9.81SSHH202 pKa = 6.62IGSATHH208 pKa = 7.0SLLLQWNEE216 pKa = 3.39LLLRR220 pKa = 11.84TGCHH224 pKa = 6.78AIFGHH229 pKa = 6.0KK230 pKa = 9.61FWGEE234 pKa = 3.59INDD237 pKa = 4.36TIRR240 pKa = 11.84QKK242 pKa = 10.91EE243 pKa = 3.74RR244 pKa = 11.84DD245 pKa = 3.61LTGVAVAGPSTQGLRR260 pKa = 11.84DD261 pKa = 3.58EE262 pKa = 4.51VRR264 pKa = 11.84AASEE268 pKa = 4.45TFWDD272 pKa = 4.02ANWRR276 pKa = 11.84MRR278 pKa = 11.84NIRR281 pKa = 11.84GVDD284 pKa = 3.37VSLGQGTRR292 pKa = 11.84RR293 pKa = 11.84GTTANYY299 pKa = 9.7AVVISRR305 pKa = 11.84VCVEE309 pKa = 4.28LLGGSSPP316 pKa = 4.02
Molecular weight: 34.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3984402 |
45 |
4952 |
494.3 |
54.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.996 ± 0.029 | 0.986 ± 0.009 |
5.561 ± 0.02 | 6.5 ± 0.024 |
3.204 ± 0.015 | 6.865 ± 0.027 |
2.483 ± 0.013 | 4.9 ± 0.017 |
4.849 ± 0.024 | 9.053 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.203 ± 0.009 | 3.495 ± 0.016 |
7.048 ± 0.033 | 3.94 ± 0.018 |
6.252 ± 0.022 | 9.583 ± 0.039 |
6.324 ± 0.017 | 6.022 ± 0.019 |
1.346 ± 0.01 | 2.391 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
