
Cowpea polerovirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus; unclassified Polerovirus
Average proteome isoelectric point is 8.3
Get precalculated fractions of proteins
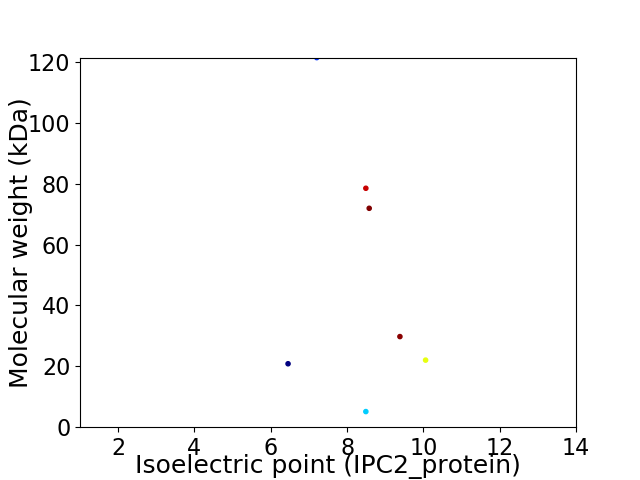
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
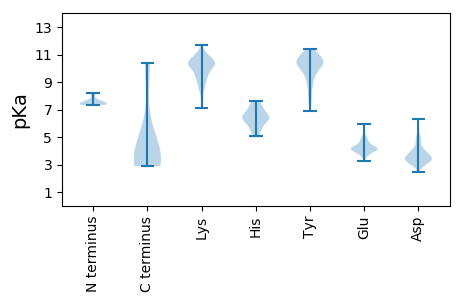
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1U9RXY8|A0A1U9RXY8_9LUTE Serine protease OS=Cowpea polerovirus 2 OX=1913125 PE=4 SV=1
MM1 pKa = 7.39EE2 pKa = 5.57GEE4 pKa = 4.73LGVADD9 pKa = 3.88AFKK12 pKa = 10.93GVSAWLWSRR21 pKa = 11.84PLGYY25 pKa = 10.23HH26 pKa = 5.52VAEE29 pKa = 4.59EE30 pKa = 4.88DD31 pKa = 3.48NDD33 pKa = 3.66EE34 pKa = 4.24TAEE37 pKa = 4.09LLQEE41 pKa = 4.23EE42 pKa = 4.71ADD44 pKa = 4.0LEE46 pKa = 4.41EE47 pKa = 4.62GRR49 pKa = 11.84ATHH52 pKa = 7.03LCFHH56 pKa = 6.7RR57 pKa = 11.84TASKK61 pKa = 10.87AVTPDD66 pKa = 2.76ISRR69 pKa = 11.84SGRR72 pKa = 11.84LFQRR76 pKa = 11.84SQSSVMEE83 pKa = 4.04FSGPTMSIRR92 pKa = 11.84SQWSSWSSSPRR103 pKa = 11.84PLQPPRR109 pKa = 11.84VQSLSNWIPIASTVPYY125 pKa = 10.27NHH127 pKa = 7.5RR128 pKa = 11.84LTNSEE133 pKa = 3.96LSKK136 pKa = 11.7GEE138 pKa = 4.14IEE140 pKa = 4.94LGTPVKK146 pKa = 10.23STGLNGTMRR155 pKa = 11.84PKK157 pKa = 10.37INSGYY162 pKa = 7.71FTRR165 pKa = 11.84EE166 pKa = 3.26TEE168 pKa = 4.11APRR171 pKa = 11.84LRR173 pKa = 11.84GPSGLPSGANSRR185 pKa = 11.84TPNRR189 pKa = 3.78
MM1 pKa = 7.39EE2 pKa = 5.57GEE4 pKa = 4.73LGVADD9 pKa = 3.88AFKK12 pKa = 10.93GVSAWLWSRR21 pKa = 11.84PLGYY25 pKa = 10.23HH26 pKa = 5.52VAEE29 pKa = 4.59EE30 pKa = 4.88DD31 pKa = 3.48NDD33 pKa = 3.66EE34 pKa = 4.24TAEE37 pKa = 4.09LLQEE41 pKa = 4.23EE42 pKa = 4.71ADD44 pKa = 4.0LEE46 pKa = 4.41EE47 pKa = 4.62GRR49 pKa = 11.84ATHH52 pKa = 7.03LCFHH56 pKa = 6.7RR57 pKa = 11.84TASKK61 pKa = 10.87AVTPDD66 pKa = 2.76ISRR69 pKa = 11.84SGRR72 pKa = 11.84LFQRR76 pKa = 11.84SQSSVMEE83 pKa = 4.04FSGPTMSIRR92 pKa = 11.84SQWSSWSSSPRR103 pKa = 11.84PLQPPRR109 pKa = 11.84VQSLSNWIPIASTVPYY125 pKa = 10.27NHH127 pKa = 7.5RR128 pKa = 11.84LTNSEE133 pKa = 3.96LSKK136 pKa = 11.7GEE138 pKa = 4.14IEE140 pKa = 4.94LGTPVKK146 pKa = 10.23STGLNGTMRR155 pKa = 11.84PKK157 pKa = 10.37INSGYY162 pKa = 7.71FTRR165 pKa = 11.84EE166 pKa = 3.26TEE168 pKa = 4.11APRR171 pKa = 11.84LRR173 pKa = 11.84GPSGLPSGANSRR185 pKa = 11.84TPNRR189 pKa = 3.78
Molecular weight: 20.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1U9RYW5|A0A1U9RYW5_9LUTE Readthrough protein OS=Cowpea polerovirus 2 OX=1913125 PE=3 SV=1
MM1 pKa = 7.44NFGNFLINEE10 pKa = 4.18QGSCVTSLKK19 pKa = 10.21RR20 pKa = 11.84LSPRR24 pKa = 11.84AVEE27 pKa = 4.21YY28 pKa = 10.46FSVLAVVCQTFLTNDD43 pKa = 3.71YY44 pKa = 9.84PSSDD48 pKa = 3.25HH49 pKa = 6.96EE50 pKa = 4.69FILRR54 pKa = 11.84CFVFLLPFLLRR65 pKa = 11.84GPKK68 pKa = 10.15NIGVRR73 pKa = 11.84KK74 pKa = 9.59PGDD77 pKa = 3.23KK78 pKa = 10.35PGLVRR83 pKa = 11.84AKK85 pKa = 9.97QFYY88 pKa = 9.7AARR91 pKa = 11.84FAARR95 pKa = 11.84LGVYY99 pKa = 8.66TPSLSGVQRR108 pKa = 11.84TVSLRR113 pKa = 11.84LSNLEE118 pKa = 3.5FTRR121 pKa = 11.84NRR123 pKa = 11.84RR124 pKa = 11.84KK125 pKa = 7.1TTAILQRR132 pKa = 11.84HH133 pKa = 5.77HH134 pKa = 6.62ARR136 pKa = 11.84SVGARR141 pKa = 11.84IEE143 pKa = 3.97RR144 pKa = 11.84RR145 pKa = 11.84KK146 pKa = 10.65DD147 pKa = 3.2VLFGGEE153 pKa = 4.06RR154 pKa = 11.84YY155 pKa = 9.4FKK157 pKa = 10.8QFVQTYY163 pKa = 10.16CRR165 pKa = 11.84FLDD168 pKa = 3.57HH169 pKa = 7.44EE170 pKa = 4.58YY171 pKa = 11.29GRR173 pKa = 11.84DD174 pKa = 3.34LSKK177 pKa = 10.87PLLVSDD183 pKa = 4.55LRR185 pKa = 11.84VVLAAAHH192 pKa = 6.72DD193 pKa = 3.97YY194 pKa = 10.5SLRR197 pKa = 11.84VAFRR201 pKa = 11.84HH202 pKa = 5.41QEE204 pKa = 3.64FHH206 pKa = 7.28ARR208 pKa = 11.84ACRR211 pKa = 11.84CLALYY216 pKa = 9.86LISSLGEE223 pKa = 3.81DD224 pKa = 3.21SALVFWVNANLPRR237 pKa = 11.84SHH239 pKa = 7.64ILFHH243 pKa = 7.54SEE245 pKa = 3.68VVLAEE250 pKa = 4.09TLLGQEE256 pKa = 4.23LQKK259 pKa = 11.27LL260 pKa = 3.88
MM1 pKa = 7.44NFGNFLINEE10 pKa = 4.18QGSCVTSLKK19 pKa = 10.21RR20 pKa = 11.84LSPRR24 pKa = 11.84AVEE27 pKa = 4.21YY28 pKa = 10.46FSVLAVVCQTFLTNDD43 pKa = 3.71YY44 pKa = 9.84PSSDD48 pKa = 3.25HH49 pKa = 6.96EE50 pKa = 4.69FILRR54 pKa = 11.84CFVFLLPFLLRR65 pKa = 11.84GPKK68 pKa = 10.15NIGVRR73 pKa = 11.84KK74 pKa = 9.59PGDD77 pKa = 3.23KK78 pKa = 10.35PGLVRR83 pKa = 11.84AKK85 pKa = 9.97QFYY88 pKa = 9.7AARR91 pKa = 11.84FAARR95 pKa = 11.84LGVYY99 pKa = 8.66TPSLSGVQRR108 pKa = 11.84TVSLRR113 pKa = 11.84LSNLEE118 pKa = 3.5FTRR121 pKa = 11.84NRR123 pKa = 11.84RR124 pKa = 11.84KK125 pKa = 7.1TTAILQRR132 pKa = 11.84HH133 pKa = 5.77HH134 pKa = 6.62ARR136 pKa = 11.84SVGARR141 pKa = 11.84IEE143 pKa = 3.97RR144 pKa = 11.84RR145 pKa = 11.84KK146 pKa = 10.65DD147 pKa = 3.2VLFGGEE153 pKa = 4.06RR154 pKa = 11.84YY155 pKa = 9.4FKK157 pKa = 10.8QFVQTYY163 pKa = 10.16CRR165 pKa = 11.84FLDD168 pKa = 3.57HH169 pKa = 7.44EE170 pKa = 4.58YY171 pKa = 11.29GRR173 pKa = 11.84DD174 pKa = 3.34LSKK177 pKa = 10.87PLLVSDD183 pKa = 4.55LRR185 pKa = 11.84VVLAAAHH192 pKa = 6.72DD193 pKa = 3.97YY194 pKa = 10.5SLRR197 pKa = 11.84VAFRR201 pKa = 11.84HH202 pKa = 5.41QEE204 pKa = 3.64FHH206 pKa = 7.28ARR208 pKa = 11.84ACRR211 pKa = 11.84CLALYY216 pKa = 9.86LISSLGEE223 pKa = 3.81DD224 pKa = 3.21SALVFWVNANLPRR237 pKa = 11.84SHH239 pKa = 7.64ILFHH243 pKa = 7.54SEE245 pKa = 3.68VVLAEE250 pKa = 4.09TLLGQEE256 pKa = 4.23LQKK259 pKa = 11.27LL260 pKa = 3.88
Molecular weight: 29.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3125 |
46 |
1080 |
446.4 |
49.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.752 ± 0.254 | 2.112 ± 0.287 |
4.096 ± 0.788 | 5.76 ± 0.579 |
4.928 ± 0.428 | 7.648 ± 0.511 |
1.76 ± 0.289 | 3.968 ± 0.507 |
5.568 ± 0.462 | 8.448 ± 1.255 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.376 ± 0.133 | 4.384 ± 0.367 |
5.76 ± 0.481 | 4.128 ± 0.392 |
7.04 ± 1.183 | 9.184 ± 0.672 |
5.184 ± 0.427 | 6.72 ± 0.565 |
2.112 ± 0.364 | 3.04 ± 0.213 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
