
Nitrosomonas communis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; Nitrosomonadaceae; Nitrosomonas
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
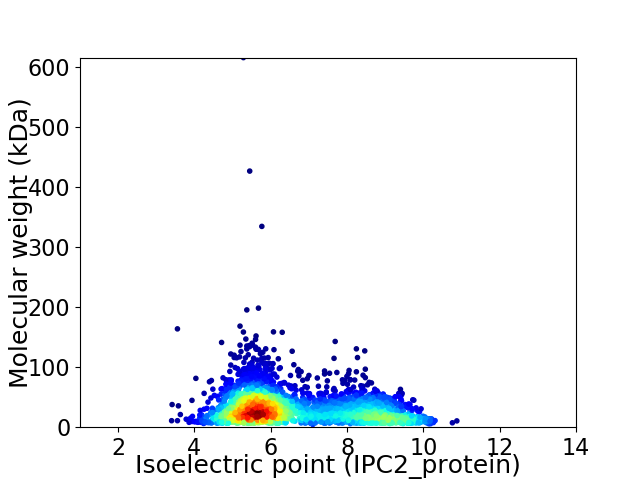
Virtual 2D-PAGE plot for 3064 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F7KJC5|A0A0F7KJC5_9PROT 23S rRNA (guanosine-2'-O-)-methyltransferase RlmB OS=Nitrosomonas communis OX=44574 GN=rlmB PE=3 SV=1
MM1 pKa = 7.22LQDD4 pKa = 4.14LNVSIIEE11 pKa = 4.31RR12 pKa = 11.84DD13 pKa = 3.34TSAGSKK19 pKa = 10.63DD20 pKa = 4.17DD21 pKa = 4.64IIFSEE26 pKa = 4.45PSNIVLNGYY35 pKa = 9.17IGNDD39 pKa = 3.5MLNGGAGNDD48 pKa = 3.73TLAGSSGNDD57 pKa = 2.88ILQGGSGRR65 pKa = 11.84DD66 pKa = 3.42LLMGGDD72 pKa = 4.05GDD74 pKa = 5.03DD75 pKa = 3.48ILRR78 pKa = 11.84GGMGEE83 pKa = 4.04NDD85 pKa = 4.21HH86 pKa = 7.18LRR88 pKa = 11.84GDD90 pKa = 4.13NGNDD94 pKa = 3.1TYY96 pKa = 11.7LFAAGDD102 pKa = 4.29GNTSIDD108 pKa = 3.76NNDD111 pKa = 3.21TGTSYY116 pKa = 11.57DD117 pKa = 3.29VLQFMEE123 pKa = 5.47GINPGEE129 pKa = 4.12VVVTRR134 pKa = 11.84DD135 pKa = 3.18SSNLYY140 pKa = 8.66LTLTSTSEE148 pKa = 4.2KK149 pKa = 9.2ITVSNYY155 pKa = 9.28FYY157 pKa = 10.77QDD159 pKa = 2.77GVGPYY164 pKa = 9.98VLDD167 pKa = 5.04AIEE170 pKa = 4.87FSDD173 pKa = 3.81GTSWDD178 pKa = 3.55VATVKK183 pKa = 10.51QKK185 pKa = 10.53VMQGTSEE192 pKa = 4.16ADD194 pKa = 3.23NLTGFASNDD203 pKa = 3.88TIDD206 pKa = 3.84GLEE209 pKa = 4.7GNDD212 pKa = 4.16SLNGAAGNDD221 pKa = 3.82TLLGGAGNDD230 pKa = 3.6TLSGSEE236 pKa = 4.3GNDD239 pKa = 2.91ILEE242 pKa = 4.97GGDD245 pKa = 3.78GTDD248 pKa = 3.06VLYY251 pKa = 11.0GGSGDD256 pKa = 3.84DD257 pKa = 3.97TLSGGAGANDD267 pKa = 4.54SLTGDD272 pKa = 4.24AGNDD276 pKa = 3.36TYY278 pKa = 11.68LFAAGEE284 pKa = 4.34GHH286 pKa = 6.34TSINNYY292 pKa = 7.73DD293 pKa = 3.75TSTSRR298 pKa = 11.84HH299 pKa = 5.37DD300 pKa = 3.36VLRR303 pKa = 11.84FLEE306 pKa = 5.26GINPGDD312 pKa = 3.58VLVSRR317 pKa = 11.84DD318 pKa = 3.45SSNLYY323 pKa = 8.64LTLAGTGEE331 pKa = 4.72KK332 pKa = 8.9ITVSSYY338 pKa = 10.47FYY340 pKa = 10.62QDD342 pKa = 2.75GAGPYY347 pKa = 9.88VLDD350 pKa = 5.45AIEE353 pKa = 4.44FSNGTSWDD361 pKa = 3.57VATVKK366 pKa = 10.64QKK368 pKa = 10.72VLQGTSGADD377 pKa = 3.16NLTGFASNDD386 pKa = 3.88TIDD389 pKa = 3.84GLEE392 pKa = 4.7GNDD395 pKa = 4.16SLNGAAGNDD404 pKa = 3.82TLLGGAGNDD413 pKa = 3.6TLSGSEE419 pKa = 4.34GNDD422 pKa = 3.2VLDD425 pKa = 5.02GGDD428 pKa = 4.09GTDD431 pKa = 3.4MLYY434 pKa = 11.04GGAGDD439 pKa = 4.13DD440 pKa = 4.17TLSGGAGTNDD450 pKa = 4.17SLTGDD455 pKa = 4.25AGNDD459 pKa = 3.36TYY461 pKa = 11.68LFAAGEE467 pKa = 4.34GHH469 pKa = 6.34TSINNYY475 pKa = 7.73DD476 pKa = 3.75TSTSRR481 pKa = 11.84HH482 pKa = 5.37DD483 pKa = 3.36VLRR486 pKa = 11.84FLEE489 pKa = 5.17GINPGEE495 pKa = 4.14VAVTRR500 pKa = 11.84NSSNLYY506 pKa = 8.77LTLTSTGEE514 pKa = 4.15KK515 pKa = 8.64ITVSNYY521 pKa = 8.98FYY523 pKa = 10.84QDD525 pKa = 2.81GVSPYY530 pKa = 10.46VLDD533 pKa = 5.43AIEE536 pKa = 4.87FSDD539 pKa = 3.81GTSWDD544 pKa = 3.55VATVKK549 pKa = 10.57QKK551 pKa = 9.17TLQGTSGADD560 pKa = 3.16NLTGFASNDD569 pKa = 3.42TMNGLAGNDD578 pKa = 3.73TLSGGNGNDD587 pKa = 3.35VLEE590 pKa = 5.25GGDD593 pKa = 4.18GTDD596 pKa = 3.12MLYY599 pKa = 10.85GGSGDD604 pKa = 3.8DD605 pKa = 3.97TLSGGAGANDD615 pKa = 4.54SLTGDD620 pKa = 4.24AGNDD624 pKa = 3.36TYY626 pKa = 11.68LFAAGEE632 pKa = 4.34GHH634 pKa = 6.34TSINNYY640 pKa = 6.6DD641 pKa = 3.79TSVGRR646 pKa = 11.84HH647 pKa = 5.02DD648 pKa = 4.07VLRR651 pKa = 11.84FMQGINPGEE660 pKa = 4.09VAVTRR665 pKa = 11.84NSSNLYY671 pKa = 8.8LTLQSTGEE679 pKa = 4.17KK680 pKa = 8.61ITINSYY686 pKa = 10.21FYY688 pKa = 10.72QDD690 pKa = 3.02GAGPYY695 pKa = 9.88VLDD698 pKa = 5.37AIEE701 pKa = 4.87FSDD704 pKa = 3.82GTSWDD709 pKa = 3.66VVTVKK714 pKa = 10.58QKK716 pKa = 9.28TLQGTSGADD725 pKa = 3.16NLTGFASNDD734 pKa = 3.42TMNGLAGNDD743 pKa = 3.73TLSGGNGNDD752 pKa = 3.35VLEE755 pKa = 5.25GGDD758 pKa = 4.18GTDD761 pKa = 3.12MLYY764 pKa = 10.85GGSGDD769 pKa = 3.8DD770 pKa = 3.97TLSGGAGTNDD780 pKa = 4.17SLTGDD785 pKa = 4.25AGNDD789 pKa = 3.36TYY791 pKa = 11.68LFAAGEE797 pKa = 4.34GHH799 pKa = 6.34TSINNYY805 pKa = 7.28DD806 pKa = 3.74TSISRR811 pKa = 11.84HH812 pKa = 5.21DD813 pKa = 3.42VLRR816 pKa = 11.84FLEE819 pKa = 5.17GINPGEE825 pKa = 4.14VAVTRR830 pKa = 11.84NSSNLYY836 pKa = 8.77LTLTSTGEE844 pKa = 4.15KK845 pKa = 8.64ITVSNYY851 pKa = 8.98FYY853 pKa = 10.84QDD855 pKa = 2.81GVSPYY860 pKa = 10.46VLDD863 pKa = 5.43AIEE866 pKa = 4.87FSDD869 pKa = 3.81GTSWDD874 pKa = 3.55VATVKK879 pKa = 10.57QKK881 pKa = 9.17TLQGTSGADD890 pKa = 3.16NLTGFASNDD899 pKa = 3.42TMNGLAGNDD908 pKa = 3.73TLSGGNGNDD917 pKa = 3.35VLEE920 pKa = 5.25GGDD923 pKa = 4.18GTDD926 pKa = 3.12MLYY929 pKa = 10.85GGSGDD934 pKa = 3.8DD935 pKa = 3.97TLSGGAGANDD945 pKa = 4.54SLTGDD950 pKa = 4.24AGNDD954 pKa = 3.36TYY956 pKa = 11.68LFAAGEE962 pKa = 4.34GHH964 pKa = 6.34TSINNYY970 pKa = 6.6DD971 pKa = 3.79TSVGRR976 pKa = 11.84HH977 pKa = 5.02DD978 pKa = 4.07VLRR981 pKa = 11.84FMQGINPGEE990 pKa = 4.09VAVTRR995 pKa = 11.84NSSNLYY1001 pKa = 8.8LTLQSTGEE1009 pKa = 4.17KK1010 pKa = 8.61ITINSYY1016 pKa = 10.21FYY1018 pKa = 10.72QDD1020 pKa = 3.02GAGPYY1025 pKa = 9.88VLDD1028 pKa = 5.37AIEE1031 pKa = 4.87FSDD1034 pKa = 3.82GTSWDD1039 pKa = 3.66VVTVKK1044 pKa = 10.58QKK1046 pKa = 9.28TLQGTSGADD1055 pKa = 3.16NLTGFASNDD1064 pKa = 3.42TMNGLAGNDD1073 pKa = 3.73TLSGGNGNDD1082 pKa = 3.35VLEE1085 pKa = 5.25GGDD1088 pKa = 4.18GTDD1091 pKa = 3.12MLYY1094 pKa = 10.85GGSGDD1099 pKa = 3.8DD1100 pKa = 3.97TLSGGAGANDD1110 pKa = 4.54SLTGDD1115 pKa = 4.24AGNDD1119 pKa = 3.36TYY1121 pKa = 11.68LFAAGEE1127 pKa = 4.64GNTSINNYY1135 pKa = 7.4DD1136 pKa = 3.65TGVGRR1141 pKa = 11.84HH1142 pKa = 5.76DD1143 pKa = 3.53VLRR1146 pKa = 11.84FLQGINPGDD1155 pKa = 3.61VLVSRR1160 pKa = 11.84DD1161 pKa = 3.45SSNLYY1166 pKa = 8.64LTLAGTGEE1174 pKa = 4.72KK1175 pKa = 8.9ITVSSYY1181 pKa = 10.47FYY1183 pKa = 10.62QDD1185 pKa = 2.75GAGPYY1190 pKa = 9.88VLDD1193 pKa = 5.37AIEE1196 pKa = 4.87FSDD1199 pKa = 3.81GTSWDD1204 pKa = 3.55VATVKK1209 pKa = 10.57QKK1211 pKa = 9.17TLQGTSGADD1220 pKa = 3.16NLTGFASNDD1229 pKa = 3.38SMNGLAGNDD1238 pKa = 3.73TLSGGNGNDD1247 pKa = 3.5VLDD1250 pKa = 4.97GGDD1253 pKa = 4.09GTDD1256 pKa = 3.4MLYY1259 pKa = 11.04GGAGDD1264 pKa = 4.13DD1265 pKa = 4.17TLSGGAGTNDD1275 pKa = 4.17SLTGDD1280 pKa = 4.25AGNDD1284 pKa = 3.36TYY1286 pKa = 11.68LFAAGEE1292 pKa = 4.64GNTSINNYY1300 pKa = 6.4DD1301 pKa = 3.7TSVGRR1306 pKa = 11.84HH1307 pKa = 5.14DD1308 pKa = 3.45VLRR1311 pKa = 11.84FLEE1314 pKa = 5.26GINPGDD1320 pKa = 3.58VLVSRR1325 pKa = 11.84DD1326 pKa = 3.45SSNLYY1331 pKa = 8.64LTLAGTGEE1339 pKa = 4.72KK1340 pKa = 8.9ITVSSYY1346 pKa = 10.47FYY1348 pKa = 10.62QDD1350 pKa = 2.75GAGPYY1355 pKa = 9.88VLDD1358 pKa = 5.37AIEE1361 pKa = 4.87FSDD1364 pKa = 3.81GTSWDD1369 pKa = 3.55VATVKK1374 pKa = 10.57QKK1376 pKa = 9.17TLQGTSGADD1385 pKa = 3.16NLTGFASNDD1394 pKa = 3.88TIDD1397 pKa = 3.84GLEE1400 pKa = 4.7GNDD1403 pKa = 4.16SLNGAAGNDD1412 pKa = 3.82TLLGGAGNDD1421 pKa = 3.33ILSGSDD1427 pKa = 3.35GNDD1430 pKa = 3.09VLEE1433 pKa = 4.99GSDD1436 pKa = 4.48GSDD1439 pKa = 2.92MLYY1442 pKa = 10.98GGAGDD1447 pKa = 3.62DD1448 pKa = 3.86TLRR1451 pKa = 11.84GGSGGNDD1458 pKa = 3.52SLTGDD1463 pKa = 4.39AGNDD1467 pKa = 3.15TYY1469 pKa = 11.53LFKK1472 pKa = 10.91ISDD1475 pKa = 3.84AKK1477 pKa = 11.06DD1478 pKa = 2.99IINNYY1483 pKa = 8.4DD1484 pKa = 3.39TDD1486 pKa = 3.86VKK1488 pKa = 11.2SFDD1491 pKa = 3.43VLRR1494 pKa = 11.84ITEE1497 pKa = 4.08VSFEE1501 pKa = 4.15NLWFSRR1507 pKa = 11.84NGSNLQINIAGTDD1520 pKa = 3.71DD1521 pKa = 3.89QLTITNWYY1529 pKa = 9.45SGNTYY1534 pKa = 10.5QLDD1537 pKa = 3.79QITADD1542 pKa = 3.3SSSLLKK1548 pKa = 10.6EE1549 pKa = 4.41HH1550 pKa = 6.94VDD1552 pKa = 3.62QLVSAMSSYY1561 pKa = 10.63KK1562 pKa = 10.39VPSGEE1567 pKa = 4.42GNIISQDD1574 pKa = 3.26VMNALQPVFNEE1585 pKa = 3.72VWLL1588 pKa = 4.14
MM1 pKa = 7.22LQDD4 pKa = 4.14LNVSIIEE11 pKa = 4.31RR12 pKa = 11.84DD13 pKa = 3.34TSAGSKK19 pKa = 10.63DD20 pKa = 4.17DD21 pKa = 4.64IIFSEE26 pKa = 4.45PSNIVLNGYY35 pKa = 9.17IGNDD39 pKa = 3.5MLNGGAGNDD48 pKa = 3.73TLAGSSGNDD57 pKa = 2.88ILQGGSGRR65 pKa = 11.84DD66 pKa = 3.42LLMGGDD72 pKa = 4.05GDD74 pKa = 5.03DD75 pKa = 3.48ILRR78 pKa = 11.84GGMGEE83 pKa = 4.04NDD85 pKa = 4.21HH86 pKa = 7.18LRR88 pKa = 11.84GDD90 pKa = 4.13NGNDD94 pKa = 3.1TYY96 pKa = 11.7LFAAGDD102 pKa = 4.29GNTSIDD108 pKa = 3.76NNDD111 pKa = 3.21TGTSYY116 pKa = 11.57DD117 pKa = 3.29VLQFMEE123 pKa = 5.47GINPGEE129 pKa = 4.12VVVTRR134 pKa = 11.84DD135 pKa = 3.18SSNLYY140 pKa = 8.66LTLTSTSEE148 pKa = 4.2KK149 pKa = 9.2ITVSNYY155 pKa = 9.28FYY157 pKa = 10.77QDD159 pKa = 2.77GVGPYY164 pKa = 9.98VLDD167 pKa = 5.04AIEE170 pKa = 4.87FSDD173 pKa = 3.81GTSWDD178 pKa = 3.55VATVKK183 pKa = 10.51QKK185 pKa = 10.53VMQGTSEE192 pKa = 4.16ADD194 pKa = 3.23NLTGFASNDD203 pKa = 3.88TIDD206 pKa = 3.84GLEE209 pKa = 4.7GNDD212 pKa = 4.16SLNGAAGNDD221 pKa = 3.82TLLGGAGNDD230 pKa = 3.6TLSGSEE236 pKa = 4.3GNDD239 pKa = 2.91ILEE242 pKa = 4.97GGDD245 pKa = 3.78GTDD248 pKa = 3.06VLYY251 pKa = 11.0GGSGDD256 pKa = 3.84DD257 pKa = 3.97TLSGGAGANDD267 pKa = 4.54SLTGDD272 pKa = 4.24AGNDD276 pKa = 3.36TYY278 pKa = 11.68LFAAGEE284 pKa = 4.34GHH286 pKa = 6.34TSINNYY292 pKa = 7.73DD293 pKa = 3.75TSTSRR298 pKa = 11.84HH299 pKa = 5.37DD300 pKa = 3.36VLRR303 pKa = 11.84FLEE306 pKa = 5.26GINPGDD312 pKa = 3.58VLVSRR317 pKa = 11.84DD318 pKa = 3.45SSNLYY323 pKa = 8.64LTLAGTGEE331 pKa = 4.72KK332 pKa = 8.9ITVSSYY338 pKa = 10.47FYY340 pKa = 10.62QDD342 pKa = 2.75GAGPYY347 pKa = 9.88VLDD350 pKa = 5.45AIEE353 pKa = 4.44FSNGTSWDD361 pKa = 3.57VATVKK366 pKa = 10.64QKK368 pKa = 10.72VLQGTSGADD377 pKa = 3.16NLTGFASNDD386 pKa = 3.88TIDD389 pKa = 3.84GLEE392 pKa = 4.7GNDD395 pKa = 4.16SLNGAAGNDD404 pKa = 3.82TLLGGAGNDD413 pKa = 3.6TLSGSEE419 pKa = 4.34GNDD422 pKa = 3.2VLDD425 pKa = 5.02GGDD428 pKa = 4.09GTDD431 pKa = 3.4MLYY434 pKa = 11.04GGAGDD439 pKa = 4.13DD440 pKa = 4.17TLSGGAGTNDD450 pKa = 4.17SLTGDD455 pKa = 4.25AGNDD459 pKa = 3.36TYY461 pKa = 11.68LFAAGEE467 pKa = 4.34GHH469 pKa = 6.34TSINNYY475 pKa = 7.73DD476 pKa = 3.75TSTSRR481 pKa = 11.84HH482 pKa = 5.37DD483 pKa = 3.36VLRR486 pKa = 11.84FLEE489 pKa = 5.17GINPGEE495 pKa = 4.14VAVTRR500 pKa = 11.84NSSNLYY506 pKa = 8.77LTLTSTGEE514 pKa = 4.15KK515 pKa = 8.64ITVSNYY521 pKa = 8.98FYY523 pKa = 10.84QDD525 pKa = 2.81GVSPYY530 pKa = 10.46VLDD533 pKa = 5.43AIEE536 pKa = 4.87FSDD539 pKa = 3.81GTSWDD544 pKa = 3.55VATVKK549 pKa = 10.57QKK551 pKa = 9.17TLQGTSGADD560 pKa = 3.16NLTGFASNDD569 pKa = 3.42TMNGLAGNDD578 pKa = 3.73TLSGGNGNDD587 pKa = 3.35VLEE590 pKa = 5.25GGDD593 pKa = 4.18GTDD596 pKa = 3.12MLYY599 pKa = 10.85GGSGDD604 pKa = 3.8DD605 pKa = 3.97TLSGGAGANDD615 pKa = 4.54SLTGDD620 pKa = 4.24AGNDD624 pKa = 3.36TYY626 pKa = 11.68LFAAGEE632 pKa = 4.34GHH634 pKa = 6.34TSINNYY640 pKa = 6.6DD641 pKa = 3.79TSVGRR646 pKa = 11.84HH647 pKa = 5.02DD648 pKa = 4.07VLRR651 pKa = 11.84FMQGINPGEE660 pKa = 4.09VAVTRR665 pKa = 11.84NSSNLYY671 pKa = 8.8LTLQSTGEE679 pKa = 4.17KK680 pKa = 8.61ITINSYY686 pKa = 10.21FYY688 pKa = 10.72QDD690 pKa = 3.02GAGPYY695 pKa = 9.88VLDD698 pKa = 5.37AIEE701 pKa = 4.87FSDD704 pKa = 3.82GTSWDD709 pKa = 3.66VVTVKK714 pKa = 10.58QKK716 pKa = 9.28TLQGTSGADD725 pKa = 3.16NLTGFASNDD734 pKa = 3.42TMNGLAGNDD743 pKa = 3.73TLSGGNGNDD752 pKa = 3.35VLEE755 pKa = 5.25GGDD758 pKa = 4.18GTDD761 pKa = 3.12MLYY764 pKa = 10.85GGSGDD769 pKa = 3.8DD770 pKa = 3.97TLSGGAGTNDD780 pKa = 4.17SLTGDD785 pKa = 4.25AGNDD789 pKa = 3.36TYY791 pKa = 11.68LFAAGEE797 pKa = 4.34GHH799 pKa = 6.34TSINNYY805 pKa = 7.28DD806 pKa = 3.74TSISRR811 pKa = 11.84HH812 pKa = 5.21DD813 pKa = 3.42VLRR816 pKa = 11.84FLEE819 pKa = 5.17GINPGEE825 pKa = 4.14VAVTRR830 pKa = 11.84NSSNLYY836 pKa = 8.77LTLTSTGEE844 pKa = 4.15KK845 pKa = 8.64ITVSNYY851 pKa = 8.98FYY853 pKa = 10.84QDD855 pKa = 2.81GVSPYY860 pKa = 10.46VLDD863 pKa = 5.43AIEE866 pKa = 4.87FSDD869 pKa = 3.81GTSWDD874 pKa = 3.55VATVKK879 pKa = 10.57QKK881 pKa = 9.17TLQGTSGADD890 pKa = 3.16NLTGFASNDD899 pKa = 3.42TMNGLAGNDD908 pKa = 3.73TLSGGNGNDD917 pKa = 3.35VLEE920 pKa = 5.25GGDD923 pKa = 4.18GTDD926 pKa = 3.12MLYY929 pKa = 10.85GGSGDD934 pKa = 3.8DD935 pKa = 3.97TLSGGAGANDD945 pKa = 4.54SLTGDD950 pKa = 4.24AGNDD954 pKa = 3.36TYY956 pKa = 11.68LFAAGEE962 pKa = 4.34GHH964 pKa = 6.34TSINNYY970 pKa = 6.6DD971 pKa = 3.79TSVGRR976 pKa = 11.84HH977 pKa = 5.02DD978 pKa = 4.07VLRR981 pKa = 11.84FMQGINPGEE990 pKa = 4.09VAVTRR995 pKa = 11.84NSSNLYY1001 pKa = 8.8LTLQSTGEE1009 pKa = 4.17KK1010 pKa = 8.61ITINSYY1016 pKa = 10.21FYY1018 pKa = 10.72QDD1020 pKa = 3.02GAGPYY1025 pKa = 9.88VLDD1028 pKa = 5.37AIEE1031 pKa = 4.87FSDD1034 pKa = 3.82GTSWDD1039 pKa = 3.66VVTVKK1044 pKa = 10.58QKK1046 pKa = 9.28TLQGTSGADD1055 pKa = 3.16NLTGFASNDD1064 pKa = 3.42TMNGLAGNDD1073 pKa = 3.73TLSGGNGNDD1082 pKa = 3.35VLEE1085 pKa = 5.25GGDD1088 pKa = 4.18GTDD1091 pKa = 3.12MLYY1094 pKa = 10.85GGSGDD1099 pKa = 3.8DD1100 pKa = 3.97TLSGGAGANDD1110 pKa = 4.54SLTGDD1115 pKa = 4.24AGNDD1119 pKa = 3.36TYY1121 pKa = 11.68LFAAGEE1127 pKa = 4.64GNTSINNYY1135 pKa = 7.4DD1136 pKa = 3.65TGVGRR1141 pKa = 11.84HH1142 pKa = 5.76DD1143 pKa = 3.53VLRR1146 pKa = 11.84FLQGINPGDD1155 pKa = 3.61VLVSRR1160 pKa = 11.84DD1161 pKa = 3.45SSNLYY1166 pKa = 8.64LTLAGTGEE1174 pKa = 4.72KK1175 pKa = 8.9ITVSSYY1181 pKa = 10.47FYY1183 pKa = 10.62QDD1185 pKa = 2.75GAGPYY1190 pKa = 9.88VLDD1193 pKa = 5.37AIEE1196 pKa = 4.87FSDD1199 pKa = 3.81GTSWDD1204 pKa = 3.55VATVKK1209 pKa = 10.57QKK1211 pKa = 9.17TLQGTSGADD1220 pKa = 3.16NLTGFASNDD1229 pKa = 3.38SMNGLAGNDD1238 pKa = 3.73TLSGGNGNDD1247 pKa = 3.5VLDD1250 pKa = 4.97GGDD1253 pKa = 4.09GTDD1256 pKa = 3.4MLYY1259 pKa = 11.04GGAGDD1264 pKa = 4.13DD1265 pKa = 4.17TLSGGAGTNDD1275 pKa = 4.17SLTGDD1280 pKa = 4.25AGNDD1284 pKa = 3.36TYY1286 pKa = 11.68LFAAGEE1292 pKa = 4.64GNTSINNYY1300 pKa = 6.4DD1301 pKa = 3.7TSVGRR1306 pKa = 11.84HH1307 pKa = 5.14DD1308 pKa = 3.45VLRR1311 pKa = 11.84FLEE1314 pKa = 5.26GINPGDD1320 pKa = 3.58VLVSRR1325 pKa = 11.84DD1326 pKa = 3.45SSNLYY1331 pKa = 8.64LTLAGTGEE1339 pKa = 4.72KK1340 pKa = 8.9ITVSSYY1346 pKa = 10.47FYY1348 pKa = 10.62QDD1350 pKa = 2.75GAGPYY1355 pKa = 9.88VLDD1358 pKa = 5.37AIEE1361 pKa = 4.87FSDD1364 pKa = 3.81GTSWDD1369 pKa = 3.55VATVKK1374 pKa = 10.57QKK1376 pKa = 9.17TLQGTSGADD1385 pKa = 3.16NLTGFASNDD1394 pKa = 3.88TIDD1397 pKa = 3.84GLEE1400 pKa = 4.7GNDD1403 pKa = 4.16SLNGAAGNDD1412 pKa = 3.82TLLGGAGNDD1421 pKa = 3.33ILSGSDD1427 pKa = 3.35GNDD1430 pKa = 3.09VLEE1433 pKa = 4.99GSDD1436 pKa = 4.48GSDD1439 pKa = 2.92MLYY1442 pKa = 10.98GGAGDD1447 pKa = 3.62DD1448 pKa = 3.86TLRR1451 pKa = 11.84GGSGGNDD1458 pKa = 3.52SLTGDD1463 pKa = 4.39AGNDD1467 pKa = 3.15TYY1469 pKa = 11.53LFKK1472 pKa = 10.91ISDD1475 pKa = 3.84AKK1477 pKa = 11.06DD1478 pKa = 2.99IINNYY1483 pKa = 8.4DD1484 pKa = 3.39TDD1486 pKa = 3.86VKK1488 pKa = 11.2SFDD1491 pKa = 3.43VLRR1494 pKa = 11.84ITEE1497 pKa = 4.08VSFEE1501 pKa = 4.15NLWFSRR1507 pKa = 11.84NGSNLQINIAGTDD1520 pKa = 3.71DD1521 pKa = 3.89QLTITNWYY1529 pKa = 9.45SGNTYY1534 pKa = 10.5QLDD1537 pKa = 3.79QITADD1542 pKa = 3.3SSSLLKK1548 pKa = 10.6EE1549 pKa = 4.41HH1550 pKa = 6.94VDD1552 pKa = 3.62QLVSAMSSYY1561 pKa = 10.63KK1562 pKa = 10.39VPSGEE1567 pKa = 4.42GNIISQDD1574 pKa = 3.26VMNALQPVFNEE1585 pKa = 3.72VWLL1588 pKa = 4.14
Molecular weight: 163.64 kDa
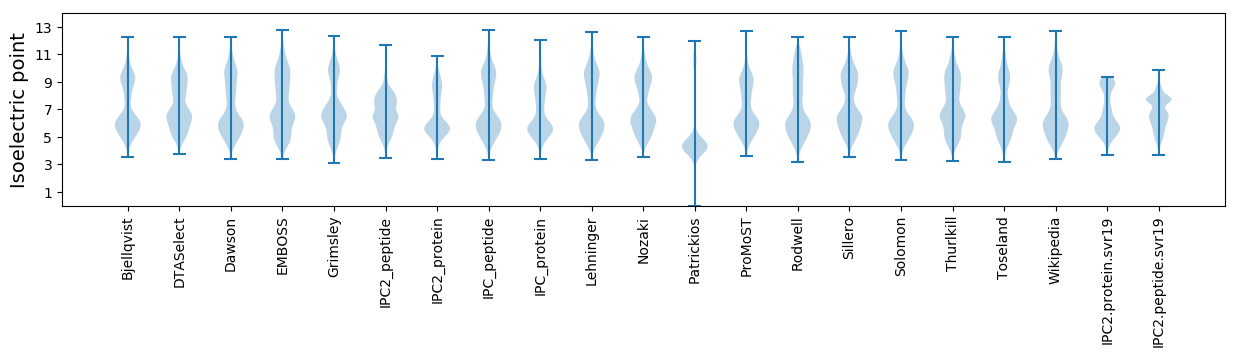
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F7KBS1|A0A0F7KBS1_9PROT Outer membrane protein assembly factor BamA OS=Nitrosomonas communis OX=44574 GN=bamA PE=3 SV=1
MM1 pKa = 7.87DD2 pKa = 5.58ASLISILLGSFTGIVLALTGAGGTIIAVPLLMFSLDD38 pKa = 3.53LTVAQAAPISLLAVSLSGAIGAWLAFRR65 pKa = 11.84QKK67 pKa = 10.09RR68 pKa = 11.84VRR70 pKa = 11.84YY71 pKa = 8.73RR72 pKa = 11.84AASFIALIGALISPAGIWLAQHH94 pKa = 6.71LPDD97 pKa = 5.28ISLTLLFAGVLLFVAQHH114 pKa = 5.99MFRR117 pKa = 11.84QSMHH121 pKa = 5.93EE122 pKa = 4.35GKK124 pKa = 10.08NEE126 pKa = 3.49MSTQHH131 pKa = 6.24SASPCQIDD139 pKa = 3.9EE140 pKa = 4.28ASGRR144 pKa = 11.84LIWTIPCFRR153 pKa = 11.84ALALSGTTAGFLSGLLGVGGGFVIVPALRR182 pKa = 11.84KK183 pKa = 8.46ITNLPILSIMATSLAITALIATTGVISAIIIRR215 pKa = 11.84AMQWPIALFFCTGTIAGMLLGRR237 pKa = 11.84MFAEE241 pKa = 4.32RR242 pKa = 11.84LAGPRR247 pKa = 11.84LQQGFAILAGGIAAGMITKK266 pKa = 8.61VIWAILLL273 pKa = 3.89
MM1 pKa = 7.87DD2 pKa = 5.58ASLISILLGSFTGIVLALTGAGGTIIAVPLLMFSLDD38 pKa = 3.53LTVAQAAPISLLAVSLSGAIGAWLAFRR65 pKa = 11.84QKK67 pKa = 10.09RR68 pKa = 11.84VRR70 pKa = 11.84YY71 pKa = 8.73RR72 pKa = 11.84AASFIALIGALISPAGIWLAQHH94 pKa = 6.71LPDD97 pKa = 5.28ISLTLLFAGVLLFVAQHH114 pKa = 5.99MFRR117 pKa = 11.84QSMHH121 pKa = 5.93EE122 pKa = 4.35GKK124 pKa = 10.08NEE126 pKa = 3.49MSTQHH131 pKa = 6.24SASPCQIDD139 pKa = 3.9EE140 pKa = 4.28ASGRR144 pKa = 11.84LIWTIPCFRR153 pKa = 11.84ALALSGTTAGFLSGLLGVGGGFVIVPALRR182 pKa = 11.84KK183 pKa = 8.46ITNLPILSIMATSLAITALIATTGVISAIIIRR215 pKa = 11.84AMQWPIALFFCTGTIAGMLLGRR237 pKa = 11.84MFAEE241 pKa = 4.32RR242 pKa = 11.84LAGPRR247 pKa = 11.84LQQGFAILAGGIAAGMITKK266 pKa = 8.61VIWAILLL273 pKa = 3.89
Molecular weight: 28.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
915887 |
42 |
5495 |
298.9 |
33.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.953 ± 0.043 | 1.047 ± 0.017 |
5.059 ± 0.032 | 6.147 ± 0.04 |
4.024 ± 0.029 | 6.812 ± 0.049 |
2.608 ± 0.02 | 6.893 ± 0.04 |
4.866 ± 0.05 | 10.658 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.507 ± 0.022 | 3.954 ± 0.035 |
4.447 ± 0.03 | 4.32 ± 0.034 |
5.668 ± 0.039 | 5.973 ± 0.036 |
5.267 ± 0.036 | 6.411 ± 0.042 |
1.383 ± 0.018 | 3.003 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
