
Spinach latent virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Ilarvirus
Average proteome isoelectric point is 7.59
Get precalculated fractions of proteins
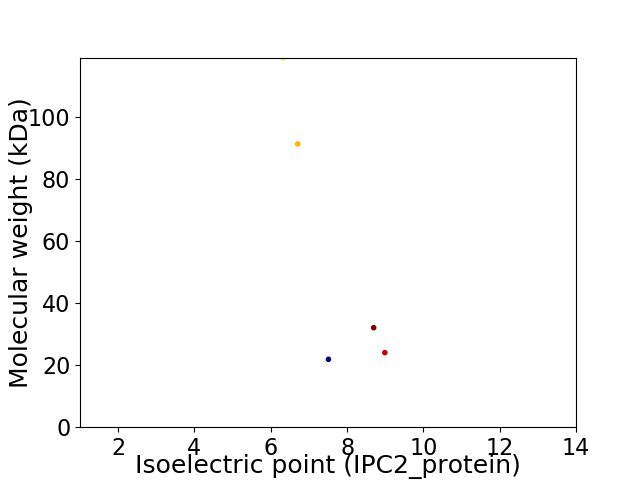
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|O57162|O57162_9BROM RNA-directed RNA polymerase 2a OS=Spinach latent virus OX=42680 PE=4 SV=1
MM1 pKa = 7.46DD2 pKa = 5.89TIVQMKK8 pKa = 7.34VLCYY12 pKa = 10.6EE13 pKa = 4.13SMILLMIPSDD23 pKa = 3.97VNAPRR28 pKa = 11.84PEE30 pKa = 3.95TAIGRR35 pKa = 11.84LISDD39 pKa = 4.03AAVSGVLRR47 pKa = 11.84QCDD50 pKa = 3.79FTAAEE55 pKa = 4.35ASQCFVPIITGRR67 pKa = 11.84SEE69 pKa = 4.38CIEE72 pKa = 3.91TRR74 pKa = 11.84FSGRR78 pKa = 11.84EE79 pKa = 3.36IQFRR83 pKa = 11.84NFSYY87 pKa = 10.71SSQSFAAAHH96 pKa = 5.94RR97 pKa = 11.84VCEE100 pKa = 3.53ADD102 pKa = 3.43YY103 pKa = 10.5IYY105 pKa = 11.24SRR107 pKa = 11.84FQTRR111 pKa = 11.84NTTIIDD117 pKa = 3.13IGGNFCTHH125 pKa = 5.67SKK127 pKa = 9.27MGRR130 pKa = 11.84DD131 pKa = 3.84NVHH134 pKa = 5.96SCCPILDD141 pKa = 3.72VRR143 pKa = 11.84DD144 pKa = 3.5GARR147 pKa = 11.84YY148 pKa = 7.17TDD150 pKa = 3.1RR151 pKa = 11.84FMSVAGALEE160 pKa = 4.11TQPDD164 pKa = 3.5RR165 pKa = 11.84EE166 pKa = 4.29LRR168 pKa = 11.84LNYY171 pKa = 9.89CSNKK175 pKa = 9.76FEE177 pKa = 5.1DD178 pKa = 5.03CDD180 pKa = 4.59VSAPWAMAIHH190 pKa = 7.06SISDD194 pKa = 3.36IPITSVVKK202 pKa = 9.02HH203 pKa = 5.74CFRR206 pKa = 11.84RR207 pKa = 11.84GVKK210 pKa = 10.07KK211 pKa = 10.81LIASVMMDD219 pKa = 3.07PMMLVASQGFMPRR232 pKa = 11.84LNVQWEE238 pKa = 4.16IEE240 pKa = 3.85PDD242 pKa = 3.56EE243 pKa = 4.3EE244 pKa = 4.53NKK246 pKa = 10.11GSRR249 pKa = 11.84RR250 pKa = 11.84ISFHH254 pKa = 6.68FVDD257 pKa = 5.61APGLSYY263 pKa = 9.32THH265 pKa = 6.8NYY267 pKa = 9.12DD268 pKa = 3.86VLMQYY273 pKa = 7.39MTCNQVIISGKK284 pKa = 8.37AAYY287 pKa = 9.46RR288 pKa = 11.84VEE290 pKa = 4.2RR291 pKa = 11.84VADD294 pKa = 3.92LSGVFIVEE302 pKa = 4.08INPGGNCWNEE312 pKa = 3.9RR313 pKa = 11.84LDD315 pKa = 4.25LVPMRR320 pKa = 11.84DD321 pKa = 3.75VSCAWMTSLKK331 pKa = 10.42RR332 pKa = 11.84KK333 pKa = 7.29TLVRR337 pKa = 11.84VAIPQYY343 pKa = 8.21IHH345 pKa = 5.77SWEE348 pKa = 4.18VKK350 pKa = 10.13YY351 pKa = 11.11VIADD355 pKa = 3.34TDD357 pKa = 3.65FVRR360 pKa = 11.84RR361 pKa = 11.84VAEE364 pKa = 3.96VSFRR368 pKa = 11.84QYY370 pKa = 11.05KK371 pKa = 10.35PEE373 pKa = 4.09TPLQDD378 pKa = 3.45LVKK381 pKa = 10.61SVATMISSASNHH393 pKa = 6.21CIINGVTMQTGTPLAIEE410 pKa = 5.36DD411 pKa = 4.34YY412 pKa = 11.36VPMAVTFVAFAKK424 pKa = 10.13SRR426 pKa = 11.84YY427 pKa = 9.52DD428 pKa = 3.62SIKK431 pKa = 10.46EE432 pKa = 3.79GMKK435 pKa = 9.35MVRR438 pKa = 11.84QRR440 pKa = 11.84GTNIADD446 pKa = 3.89PNDD449 pKa = 3.54HH450 pKa = 6.89YY451 pKa = 11.46DD452 pKa = 3.65YY453 pKa = 10.98EE454 pKa = 5.63KK455 pKa = 11.06DD456 pKa = 3.51GSDD459 pKa = 3.07VKK461 pKa = 11.14SFSNHH466 pKa = 6.48IIPMKK471 pKa = 9.99NTIKK475 pKa = 10.9SLFFPMKK482 pKa = 10.59VKK484 pKa = 10.6MNDD487 pKa = 2.91NSMLIYY493 pKa = 10.44SQGILSTVVDD503 pKa = 4.88EE504 pKa = 4.92IKK506 pKa = 9.91TLFGWDD512 pKa = 2.89VWDD515 pKa = 4.04TDD517 pKa = 3.84DD518 pKa = 6.54AVIQSLPSFYY528 pKa = 11.11KK529 pKa = 9.97MEE531 pKa = 4.99DD532 pKa = 3.35VFQVTSDD539 pKa = 4.15HH540 pKa = 6.33YY541 pKa = 10.73CLSHH545 pKa = 6.15TLSVDD550 pKa = 2.39WWLEE554 pKa = 3.65GLYY557 pKa = 10.53DD558 pKa = 3.82SYY560 pKa = 11.88HH561 pKa = 7.01DD562 pKa = 3.62MRR564 pKa = 11.84VAHH567 pKa = 6.72KK568 pKa = 10.41KK569 pKa = 10.51KK570 pKa = 10.39LDD572 pKa = 3.57EE573 pKa = 4.4EE574 pKa = 4.54EE575 pKa = 4.06SRR577 pKa = 11.84KK578 pKa = 8.37TKK580 pKa = 10.7VEE582 pKa = 3.76NALLKK587 pKa = 10.24IAEE590 pKa = 4.38VLEE593 pKa = 4.54KK594 pKa = 10.5PDD596 pKa = 3.66VSDD599 pKa = 3.56GLKK602 pKa = 10.54ALSEE606 pKa = 4.12LPIISSLLEE615 pKa = 4.11KK616 pKa = 10.47KK617 pKa = 10.48SEE619 pKa = 4.29EE620 pKa = 4.14IVTKK624 pKa = 8.01PQCRR628 pKa = 11.84DD629 pKa = 2.99TEE631 pKa = 4.24KK632 pKa = 10.9PHH634 pKa = 6.37INPYY638 pKa = 10.75ADD640 pKa = 4.6AIKK643 pKa = 9.99EE644 pKa = 3.9AQAYY648 pKa = 8.93YY649 pKa = 10.94EE650 pKa = 4.04EE651 pKa = 5.67LEE653 pKa = 4.53VVNTRR658 pKa = 11.84NLRR661 pKa = 11.84GVGDD665 pKa = 3.49YY666 pKa = 10.85LFWRR670 pKa = 11.84KK671 pKa = 9.48KK672 pKa = 10.32SNYY675 pKa = 8.75ASVWGADD682 pKa = 3.46EE683 pKa = 4.25SRR685 pKa = 11.84VVLEE689 pKa = 3.47PMARR693 pKa = 11.84KK694 pKa = 9.55FYY696 pKa = 11.01SRR698 pKa = 11.84DD699 pKa = 3.43RR700 pKa = 11.84NVSIPEE706 pKa = 4.01YY707 pKa = 9.32EE708 pKa = 4.07RR709 pKa = 11.84GMTVDD714 pKa = 4.15GFVSIKK720 pKa = 9.94WVDD723 pKa = 3.48GNITEE728 pKa = 4.7DD729 pKa = 2.89TWKK732 pKa = 10.6CLSKK736 pKa = 10.95YY737 pKa = 10.72AVVLLDD743 pKa = 3.48SSCVFDD749 pKa = 4.06AGSRR753 pKa = 11.84IMPGLKK759 pKa = 9.69KK760 pKa = 10.62ALVMDD765 pKa = 4.95ANFKK769 pKa = 10.43IVIEE773 pKa = 4.87DD774 pKa = 3.86GVAGCGKK781 pKa = 6.92TTSLLKK787 pKa = 9.87QAKK790 pKa = 8.93PDD792 pKa = 3.73SDD794 pKa = 4.45LLLAANRR801 pKa = 11.84EE802 pKa = 4.15TANDD806 pKa = 3.57AKK808 pKa = 11.0ASGVIPKK815 pKa = 10.09ALEE818 pKa = 3.76YY819 pKa = 10.19RR820 pKa = 11.84VRR822 pKa = 11.84TVDD825 pKa = 3.05SYY827 pKa = 12.31LMLRR831 pKa = 11.84TWFTAEE837 pKa = 3.69RR838 pKa = 11.84LLVDD842 pKa = 3.38EE843 pKa = 5.37CFLVHH848 pKa = 7.28AGLVYY853 pKa = 10.47AAATLARR860 pKa = 11.84VKK862 pKa = 10.46EE863 pKa = 4.43VIAFGDD869 pKa = 3.67TKK871 pKa = 10.74QIPFVSRR878 pKa = 11.84IPTVKK883 pKa = 9.94LRR885 pKa = 11.84HH886 pKa = 5.69ASVVGTLKK894 pKa = 10.55PRR896 pKa = 11.84TITYY900 pKa = 8.44RR901 pKa = 11.84CPRR904 pKa = 11.84DD905 pKa = 3.4VTAVLSEE912 pKa = 4.12KK913 pKa = 10.74FYY915 pKa = 9.14GTKK918 pKa = 10.35VKK920 pKa = 9.79TFNLVKK926 pKa = 10.61QSLEE930 pKa = 3.95LKK932 pKa = 10.49NIDD935 pKa = 3.74SSTEE939 pKa = 3.66IPVEE943 pKa = 3.97KK944 pKa = 9.58DD945 pKa = 2.83TLYY948 pKa = 10.43IAHH951 pKa = 6.15TQADD955 pKa = 3.55KK956 pKa = 10.81HH957 pKa = 7.55ALMRR961 pKa = 11.84LPGMNGIEE969 pKa = 3.97VLTTHH974 pKa = 6.78EE975 pKa = 4.37AQGKK979 pKa = 7.0TRR981 pKa = 11.84DD982 pKa = 3.52HH983 pKa = 6.77VILVRR988 pKa = 11.84LSKK991 pKa = 8.17TTNLLYY997 pKa = 10.44SGKK1000 pKa = 9.12MPDD1003 pKa = 3.7AGGSHH1008 pKa = 6.45NLVGLSRR1015 pKa = 11.84HH1016 pKa = 5.77KK1017 pKa = 10.69KK1018 pKa = 7.05SLRR1021 pKa = 11.84YY1022 pKa = 9.33FSVFADD1028 pKa = 4.69DD1029 pKa = 5.34PDD1031 pKa = 3.92DD1032 pKa = 4.48QIATGIRR1039 pKa = 11.84WSKK1042 pKa = 9.12TLDD1045 pKa = 3.57EE1046 pKa = 4.68QEE1048 pKa = 3.99LAAYY1052 pKa = 8.95RR1053 pKa = 11.84AAGG1056 pKa = 3.32
MM1 pKa = 7.46DD2 pKa = 5.89TIVQMKK8 pKa = 7.34VLCYY12 pKa = 10.6EE13 pKa = 4.13SMILLMIPSDD23 pKa = 3.97VNAPRR28 pKa = 11.84PEE30 pKa = 3.95TAIGRR35 pKa = 11.84LISDD39 pKa = 4.03AAVSGVLRR47 pKa = 11.84QCDD50 pKa = 3.79FTAAEE55 pKa = 4.35ASQCFVPIITGRR67 pKa = 11.84SEE69 pKa = 4.38CIEE72 pKa = 3.91TRR74 pKa = 11.84FSGRR78 pKa = 11.84EE79 pKa = 3.36IQFRR83 pKa = 11.84NFSYY87 pKa = 10.71SSQSFAAAHH96 pKa = 5.94RR97 pKa = 11.84VCEE100 pKa = 3.53ADD102 pKa = 3.43YY103 pKa = 10.5IYY105 pKa = 11.24SRR107 pKa = 11.84FQTRR111 pKa = 11.84NTTIIDD117 pKa = 3.13IGGNFCTHH125 pKa = 5.67SKK127 pKa = 9.27MGRR130 pKa = 11.84DD131 pKa = 3.84NVHH134 pKa = 5.96SCCPILDD141 pKa = 3.72VRR143 pKa = 11.84DD144 pKa = 3.5GARR147 pKa = 11.84YY148 pKa = 7.17TDD150 pKa = 3.1RR151 pKa = 11.84FMSVAGALEE160 pKa = 4.11TQPDD164 pKa = 3.5RR165 pKa = 11.84EE166 pKa = 4.29LRR168 pKa = 11.84LNYY171 pKa = 9.89CSNKK175 pKa = 9.76FEE177 pKa = 5.1DD178 pKa = 5.03CDD180 pKa = 4.59VSAPWAMAIHH190 pKa = 7.06SISDD194 pKa = 3.36IPITSVVKK202 pKa = 9.02HH203 pKa = 5.74CFRR206 pKa = 11.84RR207 pKa = 11.84GVKK210 pKa = 10.07KK211 pKa = 10.81LIASVMMDD219 pKa = 3.07PMMLVASQGFMPRR232 pKa = 11.84LNVQWEE238 pKa = 4.16IEE240 pKa = 3.85PDD242 pKa = 3.56EE243 pKa = 4.3EE244 pKa = 4.53NKK246 pKa = 10.11GSRR249 pKa = 11.84RR250 pKa = 11.84ISFHH254 pKa = 6.68FVDD257 pKa = 5.61APGLSYY263 pKa = 9.32THH265 pKa = 6.8NYY267 pKa = 9.12DD268 pKa = 3.86VLMQYY273 pKa = 7.39MTCNQVIISGKK284 pKa = 8.37AAYY287 pKa = 9.46RR288 pKa = 11.84VEE290 pKa = 4.2RR291 pKa = 11.84VADD294 pKa = 3.92LSGVFIVEE302 pKa = 4.08INPGGNCWNEE312 pKa = 3.9RR313 pKa = 11.84LDD315 pKa = 4.25LVPMRR320 pKa = 11.84DD321 pKa = 3.75VSCAWMTSLKK331 pKa = 10.42RR332 pKa = 11.84KK333 pKa = 7.29TLVRR337 pKa = 11.84VAIPQYY343 pKa = 8.21IHH345 pKa = 5.77SWEE348 pKa = 4.18VKK350 pKa = 10.13YY351 pKa = 11.11VIADD355 pKa = 3.34TDD357 pKa = 3.65FVRR360 pKa = 11.84RR361 pKa = 11.84VAEE364 pKa = 3.96VSFRR368 pKa = 11.84QYY370 pKa = 11.05KK371 pKa = 10.35PEE373 pKa = 4.09TPLQDD378 pKa = 3.45LVKK381 pKa = 10.61SVATMISSASNHH393 pKa = 6.21CIINGVTMQTGTPLAIEE410 pKa = 5.36DD411 pKa = 4.34YY412 pKa = 11.36VPMAVTFVAFAKK424 pKa = 10.13SRR426 pKa = 11.84YY427 pKa = 9.52DD428 pKa = 3.62SIKK431 pKa = 10.46EE432 pKa = 3.79GMKK435 pKa = 9.35MVRR438 pKa = 11.84QRR440 pKa = 11.84GTNIADD446 pKa = 3.89PNDD449 pKa = 3.54HH450 pKa = 6.89YY451 pKa = 11.46DD452 pKa = 3.65YY453 pKa = 10.98EE454 pKa = 5.63KK455 pKa = 11.06DD456 pKa = 3.51GSDD459 pKa = 3.07VKK461 pKa = 11.14SFSNHH466 pKa = 6.48IIPMKK471 pKa = 9.99NTIKK475 pKa = 10.9SLFFPMKK482 pKa = 10.59VKK484 pKa = 10.6MNDD487 pKa = 2.91NSMLIYY493 pKa = 10.44SQGILSTVVDD503 pKa = 4.88EE504 pKa = 4.92IKK506 pKa = 9.91TLFGWDD512 pKa = 2.89VWDD515 pKa = 4.04TDD517 pKa = 3.84DD518 pKa = 6.54AVIQSLPSFYY528 pKa = 11.11KK529 pKa = 9.97MEE531 pKa = 4.99DD532 pKa = 3.35VFQVTSDD539 pKa = 4.15HH540 pKa = 6.33YY541 pKa = 10.73CLSHH545 pKa = 6.15TLSVDD550 pKa = 2.39WWLEE554 pKa = 3.65GLYY557 pKa = 10.53DD558 pKa = 3.82SYY560 pKa = 11.88HH561 pKa = 7.01DD562 pKa = 3.62MRR564 pKa = 11.84VAHH567 pKa = 6.72KK568 pKa = 10.41KK569 pKa = 10.51KK570 pKa = 10.39LDD572 pKa = 3.57EE573 pKa = 4.4EE574 pKa = 4.54EE575 pKa = 4.06SRR577 pKa = 11.84KK578 pKa = 8.37TKK580 pKa = 10.7VEE582 pKa = 3.76NALLKK587 pKa = 10.24IAEE590 pKa = 4.38VLEE593 pKa = 4.54KK594 pKa = 10.5PDD596 pKa = 3.66VSDD599 pKa = 3.56GLKK602 pKa = 10.54ALSEE606 pKa = 4.12LPIISSLLEE615 pKa = 4.11KK616 pKa = 10.47KK617 pKa = 10.48SEE619 pKa = 4.29EE620 pKa = 4.14IVTKK624 pKa = 8.01PQCRR628 pKa = 11.84DD629 pKa = 2.99TEE631 pKa = 4.24KK632 pKa = 10.9PHH634 pKa = 6.37INPYY638 pKa = 10.75ADD640 pKa = 4.6AIKK643 pKa = 9.99EE644 pKa = 3.9AQAYY648 pKa = 8.93YY649 pKa = 10.94EE650 pKa = 4.04EE651 pKa = 5.67LEE653 pKa = 4.53VVNTRR658 pKa = 11.84NLRR661 pKa = 11.84GVGDD665 pKa = 3.49YY666 pKa = 10.85LFWRR670 pKa = 11.84KK671 pKa = 9.48KK672 pKa = 10.32SNYY675 pKa = 8.75ASVWGADD682 pKa = 3.46EE683 pKa = 4.25SRR685 pKa = 11.84VVLEE689 pKa = 3.47PMARR693 pKa = 11.84KK694 pKa = 9.55FYY696 pKa = 11.01SRR698 pKa = 11.84DD699 pKa = 3.43RR700 pKa = 11.84NVSIPEE706 pKa = 4.01YY707 pKa = 9.32EE708 pKa = 4.07RR709 pKa = 11.84GMTVDD714 pKa = 4.15GFVSIKK720 pKa = 9.94WVDD723 pKa = 3.48GNITEE728 pKa = 4.7DD729 pKa = 2.89TWKK732 pKa = 10.6CLSKK736 pKa = 10.95YY737 pKa = 10.72AVVLLDD743 pKa = 3.48SSCVFDD749 pKa = 4.06AGSRR753 pKa = 11.84IMPGLKK759 pKa = 9.69KK760 pKa = 10.62ALVMDD765 pKa = 4.95ANFKK769 pKa = 10.43IVIEE773 pKa = 4.87DD774 pKa = 3.86GVAGCGKK781 pKa = 6.92TTSLLKK787 pKa = 9.87QAKK790 pKa = 8.93PDD792 pKa = 3.73SDD794 pKa = 4.45LLLAANRR801 pKa = 11.84EE802 pKa = 4.15TANDD806 pKa = 3.57AKK808 pKa = 11.0ASGVIPKK815 pKa = 10.09ALEE818 pKa = 3.76YY819 pKa = 10.19RR820 pKa = 11.84VRR822 pKa = 11.84TVDD825 pKa = 3.05SYY827 pKa = 12.31LMLRR831 pKa = 11.84TWFTAEE837 pKa = 3.69RR838 pKa = 11.84LLVDD842 pKa = 3.38EE843 pKa = 5.37CFLVHH848 pKa = 7.28AGLVYY853 pKa = 10.47AAATLARR860 pKa = 11.84VKK862 pKa = 10.46EE863 pKa = 4.43VIAFGDD869 pKa = 3.67TKK871 pKa = 10.74QIPFVSRR878 pKa = 11.84IPTVKK883 pKa = 9.94LRR885 pKa = 11.84HH886 pKa = 5.69ASVVGTLKK894 pKa = 10.55PRR896 pKa = 11.84TITYY900 pKa = 8.44RR901 pKa = 11.84CPRR904 pKa = 11.84DD905 pKa = 3.4VTAVLSEE912 pKa = 4.12KK913 pKa = 10.74FYY915 pKa = 9.14GTKK918 pKa = 10.35VKK920 pKa = 9.79TFNLVKK926 pKa = 10.61QSLEE930 pKa = 3.95LKK932 pKa = 10.49NIDD935 pKa = 3.74SSTEE939 pKa = 3.66IPVEE943 pKa = 3.97KK944 pKa = 9.58DD945 pKa = 2.83TLYY948 pKa = 10.43IAHH951 pKa = 6.15TQADD955 pKa = 3.55KK956 pKa = 10.81HH957 pKa = 7.55ALMRR961 pKa = 11.84LPGMNGIEE969 pKa = 3.97VLTTHH974 pKa = 6.78EE975 pKa = 4.37AQGKK979 pKa = 7.0TRR981 pKa = 11.84DD982 pKa = 3.52HH983 pKa = 6.77VILVRR988 pKa = 11.84LSKK991 pKa = 8.17TTNLLYY997 pKa = 10.44SGKK1000 pKa = 9.12MPDD1003 pKa = 3.7AGGSHH1008 pKa = 6.45NLVGLSRR1015 pKa = 11.84HH1016 pKa = 5.77KK1017 pKa = 10.69KK1018 pKa = 7.05SLRR1021 pKa = 11.84YY1022 pKa = 9.33FSVFADD1028 pKa = 4.69DD1029 pKa = 5.34PDD1031 pKa = 3.92DD1032 pKa = 4.48QIATGIRR1039 pKa = 11.84WSKK1042 pKa = 9.12TLDD1045 pKa = 3.57EE1046 pKa = 4.68QEE1048 pKa = 3.99LAAYY1052 pKa = 8.95RR1053 pKa = 11.84AAGG1056 pKa = 3.32
Molecular weight: 119.12 kDa
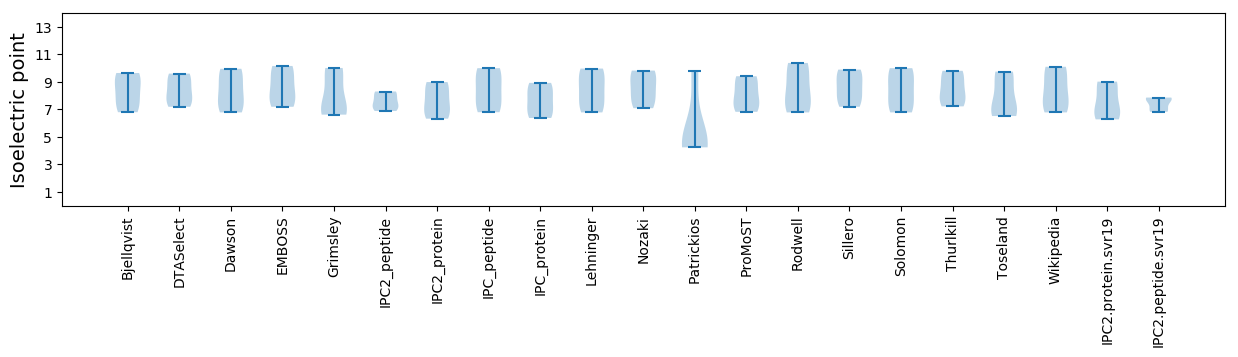
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|O57165|O57165_9BROM Putative coat protein OS=Spinach latent virus OX=42680 PE=3 SV=1
MM1 pKa = 7.53SGNAIEE7 pKa = 6.09VNGQWYY13 pKa = 10.58SPVTNNNAPSRR24 pKa = 11.84GRR26 pKa = 11.84GRR28 pKa = 11.84GRR30 pKa = 11.84GPTARR35 pKa = 11.84SRR37 pKa = 11.84NWAQSRR43 pKa = 11.84ANAARR48 pKa = 11.84SRR50 pKa = 11.84PQTLVIGTMPTNLPTWKK67 pKa = 10.16SFPGEE72 pKa = 3.75QWHH75 pKa = 5.67VVSGFSFPDD84 pKa = 2.7RR85 pKa = 11.84WNNGNIAYY93 pKa = 10.27ASMKK97 pKa = 10.17TEE99 pKa = 3.97LGKK102 pKa = 10.43IKK104 pKa = 9.39TLHH107 pKa = 6.95EE108 pKa = 3.83ITKK111 pKa = 9.57VYY113 pKa = 10.9SVMYY117 pKa = 10.15GFICKK122 pKa = 9.71SDD124 pKa = 3.16GHH126 pKa = 8.08AGFITGFDD134 pKa = 3.93TNNPTGPVAPDD145 pKa = 3.01RR146 pKa = 11.84VKK148 pKa = 10.85VKK150 pKa = 10.68KK151 pKa = 9.96NAYY154 pKa = 8.68CAKK157 pKa = 9.84QQVFPAGTTVSEE169 pKa = 5.02LKK171 pKa = 10.63NDD173 pKa = 3.14WSFIWEE179 pKa = 4.21FDD181 pKa = 3.29AAPVVGTEE189 pKa = 3.99KK190 pKa = 10.56QVTVTQFWVSTTPLPGVKK208 pKa = 9.66PPSNFLVCEE217 pKa = 4.19DD218 pKa = 3.64
MM1 pKa = 7.53SGNAIEE7 pKa = 6.09VNGQWYY13 pKa = 10.58SPVTNNNAPSRR24 pKa = 11.84GRR26 pKa = 11.84GRR28 pKa = 11.84GRR30 pKa = 11.84GPTARR35 pKa = 11.84SRR37 pKa = 11.84NWAQSRR43 pKa = 11.84ANAARR48 pKa = 11.84SRR50 pKa = 11.84PQTLVIGTMPTNLPTWKK67 pKa = 10.16SFPGEE72 pKa = 3.75QWHH75 pKa = 5.67VVSGFSFPDD84 pKa = 2.7RR85 pKa = 11.84WNNGNIAYY93 pKa = 10.27ASMKK97 pKa = 10.17TEE99 pKa = 3.97LGKK102 pKa = 10.43IKK104 pKa = 9.39TLHH107 pKa = 6.95EE108 pKa = 3.83ITKK111 pKa = 9.57VYY113 pKa = 10.9SVMYY117 pKa = 10.15GFICKK122 pKa = 9.71SDD124 pKa = 3.16GHH126 pKa = 8.08AGFITGFDD134 pKa = 3.93TNNPTGPVAPDD145 pKa = 3.01RR146 pKa = 11.84VKK148 pKa = 10.85VKK150 pKa = 10.68KK151 pKa = 9.96NAYY154 pKa = 8.68CAKK157 pKa = 9.84QQVFPAGTTVSEE169 pKa = 5.02LKK171 pKa = 10.63NDD173 pKa = 3.14WSFIWEE179 pKa = 4.21FDD181 pKa = 3.29AAPVVGTEE189 pKa = 3.99KK190 pKa = 10.56QVTVTQFWVSTTPLPGVKK208 pKa = 9.66PPSNFLVCEE217 pKa = 4.19DD218 pKa = 3.64
Molecular weight: 24.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2548 |
195 |
1056 |
509.6 |
57.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.633 ± 0.534 | 2.119 ± 0.136 |
6.436 ± 0.679 | 5.024 ± 0.376 |
4.199 ± 0.347 | 5.102 ± 0.443 |
2.433 ± 0.383 | 5.416 ± 0.538 |
6.476 ± 0.194 | 8.203 ± 0.708 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.179 ± 0.228 | 4.16 ± 0.555 |
5.102 ± 0.693 | 3.1 ± 0.234 |
5.808 ± 0.114 | 7.535 ± 0.305 |
5.495 ± 0.49 | 8.399 ± 0.191 |
1.727 ± 0.286 | 3.454 ± 0.224 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
