
Salicola phage SCTP-2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; unclassified Myoviridae
Average proteome isoelectric point is 5.91
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 541 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
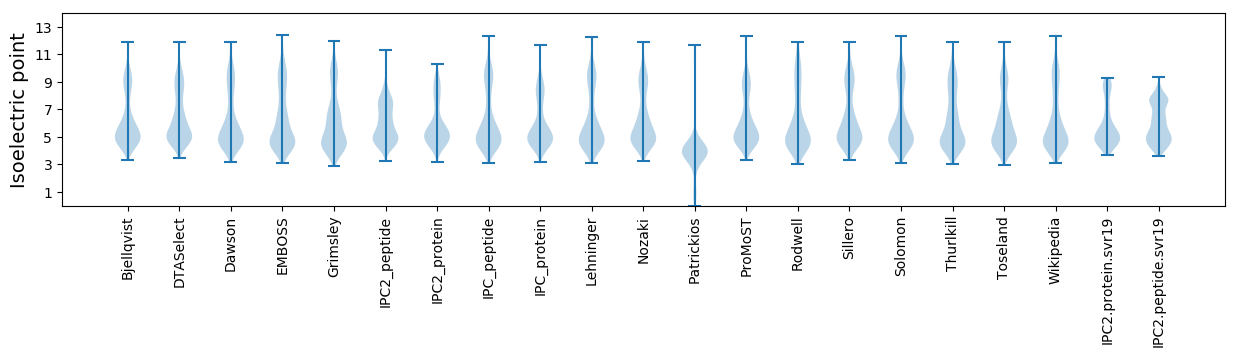
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A248SKG2|A0A248SKG2_9CAUD Uncharacterized protein OS=Salicola phage SCTP-2 OX=2015814 GN=343 PE=4 SV=1
MM1 pKa = 7.9SDD3 pKa = 3.06TKK5 pKa = 11.17NYY7 pKa = 9.37GISGIGSNVEE17 pKa = 3.51LGKK20 pKa = 10.57RR21 pKa = 11.84GNRR24 pKa = 11.84FKK26 pKa = 11.52NEE28 pKa = 3.42TDD30 pKa = 3.52KK31 pKa = 11.43ISVRR35 pKa = 11.84DD36 pKa = 3.56FDD38 pKa = 4.56DD39 pKa = 4.61ANFVPISAGDD49 pKa = 3.65PVGDD53 pKa = 3.62NDD55 pKa = 5.82LVTKK59 pKa = 10.56SYY61 pKa = 11.38LEE63 pKa = 3.9QQAFDD68 pKa = 4.16PTLKK72 pKa = 11.01SPDD75 pKa = 4.54DD76 pKa = 4.29GDD78 pKa = 3.94LTDD81 pKa = 4.91GAVTDD86 pKa = 3.75WTVGSTTYY94 pKa = 9.61STAIDD99 pKa = 4.38DD100 pKa = 4.39LNGILGRR107 pKa = 11.84LVPTAPPEE115 pKa = 4.08LSTFTINATSGSNQLLAQNAIDD137 pKa = 3.8NTTNAPSAGSSVYY150 pKa = 10.18TVFDD154 pKa = 3.4SSFQSTQANGGNGAFGGDD172 pKa = 3.45VEE174 pKa = 4.89GADD177 pKa = 3.74GNLFLSGEE185 pKa = 4.26TGTLFSYY192 pKa = 11.58VNGTQDD198 pKa = 3.19GSIVLDD204 pKa = 3.76GTDD207 pKa = 3.13NTGTYY212 pKa = 10.61GSLEE216 pKa = 4.23VLSDD220 pKa = 3.17TAYY223 pKa = 9.97PADD226 pKa = 3.64TPGFYY231 pKa = 9.31TALKK235 pKa = 10.58ARR237 pKa = 11.84INKK240 pKa = 9.17SAVSIGYY247 pKa = 8.97NEE249 pKa = 4.9AYY251 pKa = 10.34LEE253 pKa = 4.17HH254 pKa = 7.17SEE256 pKa = 4.22TGSTNTVSWVYY267 pKa = 11.45DD268 pKa = 3.73DD269 pKa = 4.04VANAPSISNQSYY281 pKa = 10.43SLDD284 pKa = 3.62SAVSKK289 pKa = 10.68QSSGINHH296 pKa = 5.63MTDD299 pKa = 2.95GDD301 pKa = 4.24TLNVSATVDD310 pKa = 3.36NLVGQTYY317 pKa = 8.04ITGTVLRR324 pKa = 11.84FTTGVGVGTLNMDD337 pKa = 4.02ATQLGTGLTFPLDD350 pKa = 3.82ANLGPQTTSGKK361 pKa = 7.52TLSIASTATFDD372 pKa = 3.42EE373 pKa = 4.11TDD375 pKa = 3.19IQVRR379 pKa = 11.84AWNANGATGSSVVSGDD395 pKa = 3.27PVLVYY400 pKa = 9.5TSALEE405 pKa = 4.46GGNKK409 pKa = 8.73PHH411 pKa = 7.2EE412 pKa = 4.31SQINGSSADD421 pKa = 3.11AYY423 pKa = 10.48RR424 pKa = 11.84YY425 pKa = 9.78YY426 pKa = 10.61LGSGFTGDD434 pKa = 3.98TPSGTLDD441 pKa = 4.41APTTTNWDD449 pKa = 3.37STQLLNASGYY459 pKa = 6.56EE460 pKa = 4.05HH461 pKa = 7.17EE462 pKa = 4.86AVTVAGVVKK471 pKa = 10.51HH472 pKa = 6.11DD473 pKa = 3.56TTNYY477 pKa = 5.21TTGYY481 pKa = 10.17IPVTTGADD489 pKa = 3.52YY490 pKa = 9.42STKK493 pKa = 10.0GTDD496 pKa = 3.1QYY498 pKa = 10.23VTYY501 pKa = 10.83VFNQSTLSSISLDD514 pKa = 3.0ISGSYY519 pKa = 9.76SGLWVALPGISDD531 pKa = 4.19DD532 pKa = 5.52AGISPNALGGNWWDD546 pKa = 4.13AFEE549 pKa = 5.28LYY551 pKa = 10.26NGSGAPGRR559 pKa = 11.84TGDD562 pKa = 3.8SSAGCAVGSAASGSSGTVNLTFGTQSTTNSTNNAVIVRR600 pKa = 11.84IKK602 pKa = 10.87LNAGDD607 pKa = 4.75SISALSIGNTPP618 pKa = 3.26
MM1 pKa = 7.9SDD3 pKa = 3.06TKK5 pKa = 11.17NYY7 pKa = 9.37GISGIGSNVEE17 pKa = 3.51LGKK20 pKa = 10.57RR21 pKa = 11.84GNRR24 pKa = 11.84FKK26 pKa = 11.52NEE28 pKa = 3.42TDD30 pKa = 3.52KK31 pKa = 11.43ISVRR35 pKa = 11.84DD36 pKa = 3.56FDD38 pKa = 4.56DD39 pKa = 4.61ANFVPISAGDD49 pKa = 3.65PVGDD53 pKa = 3.62NDD55 pKa = 5.82LVTKK59 pKa = 10.56SYY61 pKa = 11.38LEE63 pKa = 3.9QQAFDD68 pKa = 4.16PTLKK72 pKa = 11.01SPDD75 pKa = 4.54DD76 pKa = 4.29GDD78 pKa = 3.94LTDD81 pKa = 4.91GAVTDD86 pKa = 3.75WTVGSTTYY94 pKa = 9.61STAIDD99 pKa = 4.38DD100 pKa = 4.39LNGILGRR107 pKa = 11.84LVPTAPPEE115 pKa = 4.08LSTFTINATSGSNQLLAQNAIDD137 pKa = 3.8NTTNAPSAGSSVYY150 pKa = 10.18TVFDD154 pKa = 3.4SSFQSTQANGGNGAFGGDD172 pKa = 3.45VEE174 pKa = 4.89GADD177 pKa = 3.74GNLFLSGEE185 pKa = 4.26TGTLFSYY192 pKa = 11.58VNGTQDD198 pKa = 3.19GSIVLDD204 pKa = 3.76GTDD207 pKa = 3.13NTGTYY212 pKa = 10.61GSLEE216 pKa = 4.23VLSDD220 pKa = 3.17TAYY223 pKa = 9.97PADD226 pKa = 3.64TPGFYY231 pKa = 9.31TALKK235 pKa = 10.58ARR237 pKa = 11.84INKK240 pKa = 9.17SAVSIGYY247 pKa = 8.97NEE249 pKa = 4.9AYY251 pKa = 10.34LEE253 pKa = 4.17HH254 pKa = 7.17SEE256 pKa = 4.22TGSTNTVSWVYY267 pKa = 11.45DD268 pKa = 3.73DD269 pKa = 4.04VANAPSISNQSYY281 pKa = 10.43SLDD284 pKa = 3.62SAVSKK289 pKa = 10.68QSSGINHH296 pKa = 5.63MTDD299 pKa = 2.95GDD301 pKa = 4.24TLNVSATVDD310 pKa = 3.36NLVGQTYY317 pKa = 8.04ITGTVLRR324 pKa = 11.84FTTGVGVGTLNMDD337 pKa = 4.02ATQLGTGLTFPLDD350 pKa = 3.82ANLGPQTTSGKK361 pKa = 7.52TLSIASTATFDD372 pKa = 3.42EE373 pKa = 4.11TDD375 pKa = 3.19IQVRR379 pKa = 11.84AWNANGATGSSVVSGDD395 pKa = 3.27PVLVYY400 pKa = 9.5TSALEE405 pKa = 4.46GGNKK409 pKa = 8.73PHH411 pKa = 7.2EE412 pKa = 4.31SQINGSSADD421 pKa = 3.11AYY423 pKa = 10.48RR424 pKa = 11.84YY425 pKa = 9.78YY426 pKa = 10.61LGSGFTGDD434 pKa = 3.98TPSGTLDD441 pKa = 4.41APTTTNWDD449 pKa = 3.37STQLLNASGYY459 pKa = 6.56EE460 pKa = 4.05HH461 pKa = 7.17EE462 pKa = 4.86AVTVAGVVKK471 pKa = 10.51HH472 pKa = 6.11DD473 pKa = 3.56TTNYY477 pKa = 5.21TTGYY481 pKa = 10.17IPVTTGADD489 pKa = 3.52YY490 pKa = 9.42STKK493 pKa = 10.0GTDD496 pKa = 3.1QYY498 pKa = 10.23VTYY501 pKa = 10.83VFNQSTLSSISLDD514 pKa = 3.0ISGSYY519 pKa = 9.76SGLWVALPGISDD531 pKa = 4.19DD532 pKa = 5.52AGISPNALGGNWWDD546 pKa = 4.13AFEE549 pKa = 5.28LYY551 pKa = 10.26NGSGAPGRR559 pKa = 11.84TGDD562 pKa = 3.8SSAGCAVGSAASGSSGTVNLTFGTQSTTNSTNNAVIVRR600 pKa = 11.84IKK602 pKa = 10.87LNAGDD607 pKa = 4.75SISALSIGNTPP618 pKa = 3.26
Molecular weight: 63.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A248SK25|A0A248SK25_9CAUD Uncharacterized protein OS=Salicola phage SCTP-2 OX=2015814 GN=431 PE=4 SV=1
MM1 pKa = 7.7INTFKK6 pKa = 10.99CNSNEE11 pKa = 3.84WIHH14 pKa = 5.37NKK16 pKa = 9.93KK17 pKa = 9.3YY18 pKa = 10.45NSWRR22 pKa = 11.84HH23 pKa = 3.4RR24 pKa = 11.84TKK26 pKa = 10.67RR27 pKa = 11.84KK28 pKa = 9.81IIFNPILRR36 pKa = 11.84FVQGYY41 pKa = 7.8PFNKK45 pKa = 9.5RR46 pKa = 11.84PFIIASICDD55 pKa = 3.72NKK57 pKa = 10.65NGRR60 pKa = 11.84PFHH63 pKa = 6.25TGG65 pKa = 2.63
MM1 pKa = 7.7INTFKK6 pKa = 10.99CNSNEE11 pKa = 3.84WIHH14 pKa = 5.37NKK16 pKa = 9.93KK17 pKa = 9.3YY18 pKa = 10.45NSWRR22 pKa = 11.84HH23 pKa = 3.4RR24 pKa = 11.84TKK26 pKa = 10.67RR27 pKa = 11.84KK28 pKa = 9.81IIFNPILRR36 pKa = 11.84FVQGYY41 pKa = 7.8PFNKK45 pKa = 9.5RR46 pKa = 11.84PFIIASICDD55 pKa = 3.72NKK57 pKa = 10.65NGRR60 pKa = 11.84PFHH63 pKa = 6.25TGG65 pKa = 2.63
Molecular weight: 7.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
134053 |
19 |
3807 |
247.8 |
28.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.358 ± 0.095 | 1.05 ± 0.057 |
7.63 ± 0.124 | 7.256 ± 0.13 |
4.671 ± 0.107 | 4.519 ± 0.14 |
2.251 ± 0.075 | 8.658 ± 0.12 |
7.834 ± 0.21 | 7.165 ± 0.105 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.349 ± 0.086 | 8.839 ± 0.152 |
2.709 ± 0.064 | 3.5 ± 0.089 |
3.457 ± 0.088 | 7.214 ± 0.161 |
5.413 ± 0.154 | 5.407 ± 0.099 |
0.971 ± 0.043 | 5.751 ± 0.104 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
