
Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Formosa; Formosa agariphila
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
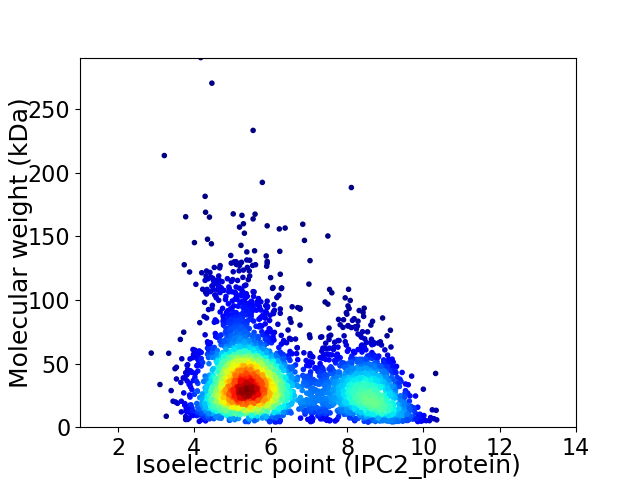
Virtual 2D-PAGE plot for 3557 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|T2KLV0|T2KLV0_FORAG Macrophage infectivity potentiator-related protein OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901) OX=1347342 GN=BN863_16890 PE=4 SV=1
MM1 pKa = 7.08ITNSTKK7 pKa = 10.73VLVLCALALSLFNCSSEE24 pKa = 4.61DD25 pKa = 3.44DD26 pKa = 3.61TDD28 pKa = 3.62RR29 pKa = 11.84GNWQEE34 pKa = 3.87RR35 pKa = 11.84SVFDD39 pKa = 3.48GVPRR43 pKa = 11.84SSVVGFVIDD52 pKa = 3.32NLGYY56 pKa = 10.05MGTGYY61 pKa = 10.9DD62 pKa = 3.68GDD64 pKa = 5.47DD65 pKa = 3.66YY66 pKa = 11.53LTDD69 pKa = 3.65FWEE72 pKa = 4.24YY73 pKa = 10.99NIEE76 pKa = 3.67GDD78 pKa = 3.85YY79 pKa = 10.02WVQKK83 pKa = 11.04ADD85 pKa = 3.7FPGTEE90 pKa = 3.88RR91 pKa = 11.84SAASGFTIGGLGYY104 pKa = 10.67LGLGYY109 pKa = 10.68DD110 pKa = 4.07GTDD113 pKa = 3.2EE114 pKa = 6.11LKK116 pKa = 11.04DD117 pKa = 3.59FWEE120 pKa = 4.45YY121 pKa = 11.66NPGSNAWTQKK131 pKa = 10.84ADD133 pKa = 3.41FLGGVRR139 pKa = 11.84QAAIGFAANNTGYY152 pKa = 10.98VGTGYY157 pKa = 10.89DD158 pKa = 3.69GDD160 pKa = 3.92NDD162 pKa = 3.77RR163 pKa = 11.84KK164 pKa = 10.76DD165 pKa = 3.62FYY167 pKa = 11.03KK168 pKa = 11.01YY169 pKa = 11.09NPTTDD174 pKa = 2.84TWSEE178 pKa = 3.74LVGFGGEE185 pKa = 3.79KK186 pKa = 9.99RR187 pKa = 11.84RR188 pKa = 11.84FATTFNIGDD197 pKa = 3.68KK198 pKa = 10.7VYY200 pKa = 10.65IGTGVSNGLYY210 pKa = 8.68KK211 pKa = 10.35TDD213 pKa = 3.62FWEE216 pKa = 4.39FDD218 pKa = 3.8PLSEE222 pKa = 4.28TFSRR226 pKa = 11.84LNDD229 pKa = 3.66LDD231 pKa = 4.99EE232 pKa = 4.97EE233 pKa = 4.75DD234 pKa = 5.58DD235 pKa = 3.96YY236 pKa = 11.87TITRR240 pKa = 11.84SNAVGFSVDD249 pKa = 2.94GLGYY253 pKa = 10.03IVSGYY258 pKa = 9.69NGSVLGSTWEE268 pKa = 4.07YY269 pKa = 11.67SPGTDD274 pKa = 2.18SWEE277 pKa = 4.24EE278 pKa = 3.54ITGIEE283 pKa = 4.31AYY285 pKa = 10.23SRR287 pKa = 11.84QDD289 pKa = 3.34ALAFSNDD296 pKa = 2.25SRR298 pKa = 11.84AFVLLGRR305 pKa = 11.84SGSLYY310 pKa = 10.73LDD312 pKa = 4.16DD313 pKa = 5.42NYY315 pKa = 11.1EE316 pKa = 4.53LFPQDD321 pKa = 5.52DD322 pKa = 4.38YY323 pKa = 12.22DD324 pKa = 6.49DD325 pKa = 4.25EE326 pKa = 4.75DD327 pKa = 3.58
MM1 pKa = 7.08ITNSTKK7 pKa = 10.73VLVLCALALSLFNCSSEE24 pKa = 4.61DD25 pKa = 3.44DD26 pKa = 3.61TDD28 pKa = 3.62RR29 pKa = 11.84GNWQEE34 pKa = 3.87RR35 pKa = 11.84SVFDD39 pKa = 3.48GVPRR43 pKa = 11.84SSVVGFVIDD52 pKa = 3.32NLGYY56 pKa = 10.05MGTGYY61 pKa = 10.9DD62 pKa = 3.68GDD64 pKa = 5.47DD65 pKa = 3.66YY66 pKa = 11.53LTDD69 pKa = 3.65FWEE72 pKa = 4.24YY73 pKa = 10.99NIEE76 pKa = 3.67GDD78 pKa = 3.85YY79 pKa = 10.02WVQKK83 pKa = 11.04ADD85 pKa = 3.7FPGTEE90 pKa = 3.88RR91 pKa = 11.84SAASGFTIGGLGYY104 pKa = 10.67LGLGYY109 pKa = 10.68DD110 pKa = 4.07GTDD113 pKa = 3.2EE114 pKa = 6.11LKK116 pKa = 11.04DD117 pKa = 3.59FWEE120 pKa = 4.45YY121 pKa = 11.66NPGSNAWTQKK131 pKa = 10.84ADD133 pKa = 3.41FLGGVRR139 pKa = 11.84QAAIGFAANNTGYY152 pKa = 10.98VGTGYY157 pKa = 10.89DD158 pKa = 3.69GDD160 pKa = 3.92NDD162 pKa = 3.77RR163 pKa = 11.84KK164 pKa = 10.76DD165 pKa = 3.62FYY167 pKa = 11.03KK168 pKa = 11.01YY169 pKa = 11.09NPTTDD174 pKa = 2.84TWSEE178 pKa = 3.74LVGFGGEE185 pKa = 3.79KK186 pKa = 9.99RR187 pKa = 11.84RR188 pKa = 11.84FATTFNIGDD197 pKa = 3.68KK198 pKa = 10.7VYY200 pKa = 10.65IGTGVSNGLYY210 pKa = 8.68KK211 pKa = 10.35TDD213 pKa = 3.62FWEE216 pKa = 4.39FDD218 pKa = 3.8PLSEE222 pKa = 4.28TFSRR226 pKa = 11.84LNDD229 pKa = 3.66LDD231 pKa = 4.99EE232 pKa = 4.97EE233 pKa = 4.75DD234 pKa = 5.58DD235 pKa = 3.96YY236 pKa = 11.87TITRR240 pKa = 11.84SNAVGFSVDD249 pKa = 2.94GLGYY253 pKa = 10.03IVSGYY258 pKa = 9.69NGSVLGSTWEE268 pKa = 4.07YY269 pKa = 11.67SPGTDD274 pKa = 2.18SWEE277 pKa = 4.24EE278 pKa = 3.54ITGIEE283 pKa = 4.31AYY285 pKa = 10.23SRR287 pKa = 11.84QDD289 pKa = 3.34ALAFSNDD296 pKa = 2.25SRR298 pKa = 11.84AFVLLGRR305 pKa = 11.84SGSLYY310 pKa = 10.73LDD312 pKa = 4.16DD313 pKa = 5.42NYY315 pKa = 11.1EE316 pKa = 4.53LFPQDD321 pKa = 5.52DD322 pKa = 4.38YY323 pKa = 12.22DD324 pKa = 6.49DD325 pKa = 4.25EE326 pKa = 4.75DD327 pKa = 3.58
Molecular weight: 36.31 kDa
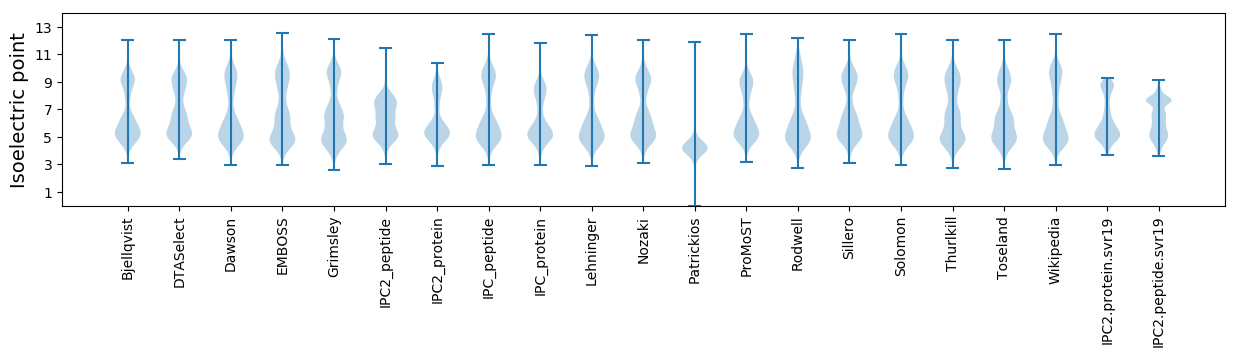
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T2KIR4|T2KIR4_FORAG Putative restriction endonuclease OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901) OX=1347342 GN=BN863_10700 PE=4 SV=1
MM1 pKa = 7.3ARR3 pKa = 11.84IAGVDD8 pKa = 3.06IPKK11 pKa = 10.02QKK13 pKa = 10.48RR14 pKa = 11.84GVISLTYY21 pKa = 9.48IYY23 pKa = 10.73GIGTSRR29 pKa = 11.84AKK31 pKa = 10.55EE32 pKa = 3.63ILAEE36 pKa = 4.21AKK38 pKa = 10.13VDD40 pKa = 3.89EE41 pKa = 4.41NTKK44 pKa = 9.59VQDD47 pKa = 3.34WTDD50 pKa = 3.58DD51 pKa = 3.56QIGSIRR57 pKa = 11.84EE58 pKa = 4.03AVSTFTIEE66 pKa = 3.9GEE68 pKa = 4.24LRR70 pKa = 11.84SEE72 pKa = 4.15TQLSIKK78 pKa = 10.3RR79 pKa = 11.84LMDD82 pKa = 3.67IGCYY86 pKa = 9.51RR87 pKa = 11.84GIRR90 pKa = 11.84HH91 pKa = 6.49RR92 pKa = 11.84AGLPLRR98 pKa = 11.84GQRR101 pKa = 11.84TKK103 pKa = 11.3NNSRR107 pKa = 11.84TRR109 pKa = 11.84KK110 pKa = 9.17GRR112 pKa = 11.84RR113 pKa = 11.84KK114 pKa = 7.81TVANKK119 pKa = 10.07KK120 pKa = 9.89KK121 pKa = 9.38ATKK124 pKa = 10.14
MM1 pKa = 7.3ARR3 pKa = 11.84IAGVDD8 pKa = 3.06IPKK11 pKa = 10.02QKK13 pKa = 10.48RR14 pKa = 11.84GVISLTYY21 pKa = 9.48IYY23 pKa = 10.73GIGTSRR29 pKa = 11.84AKK31 pKa = 10.55EE32 pKa = 3.63ILAEE36 pKa = 4.21AKK38 pKa = 10.13VDD40 pKa = 3.89EE41 pKa = 4.41NTKK44 pKa = 9.59VQDD47 pKa = 3.34WTDD50 pKa = 3.58DD51 pKa = 3.56QIGSIRR57 pKa = 11.84EE58 pKa = 4.03AVSTFTIEE66 pKa = 3.9GEE68 pKa = 4.24LRR70 pKa = 11.84SEE72 pKa = 4.15TQLSIKK78 pKa = 10.3RR79 pKa = 11.84LMDD82 pKa = 3.67IGCYY86 pKa = 9.51RR87 pKa = 11.84GIRR90 pKa = 11.84HH91 pKa = 6.49RR92 pKa = 11.84AGLPLRR98 pKa = 11.84GQRR101 pKa = 11.84TKK103 pKa = 11.3NNSRR107 pKa = 11.84TRR109 pKa = 11.84KK110 pKa = 9.17GRR112 pKa = 11.84RR113 pKa = 11.84KK114 pKa = 7.81TVANKK119 pKa = 10.07KK120 pKa = 9.89KK121 pKa = 9.38ATKK124 pKa = 10.14
Molecular weight: 13.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1231760 |
37 |
2753 |
346.3 |
39.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.39 ± 0.046 | 0.724 ± 0.012 |
5.892 ± 0.034 | 6.453 ± 0.035 |
5.133 ± 0.031 | 6.379 ± 0.042 |
1.909 ± 0.02 | 7.819 ± 0.039 |
7.467 ± 0.047 | 9.212 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.141 ± 0.023 | 6.237 ± 0.042 |
3.421 ± 0.023 | 3.349 ± 0.022 |
3.244 ± 0.025 | 6.565 ± 0.034 |
6.083 ± 0.04 | 6.287 ± 0.032 |
1.114 ± 0.015 | 4.182 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
