
Bromus-associated circular DNA virus 2
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.63
Get precalculated fractions of proteins
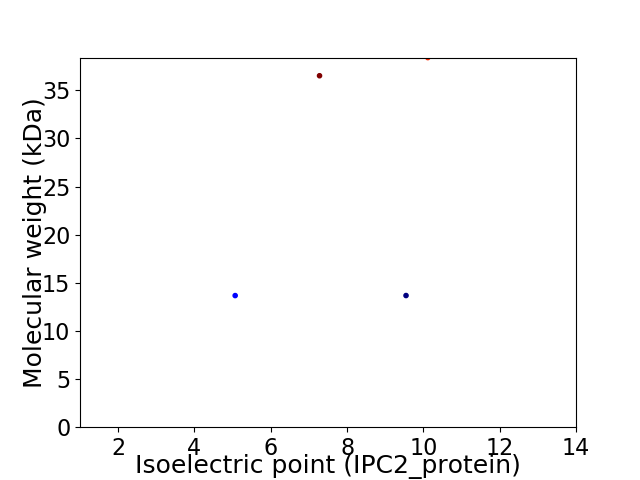
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
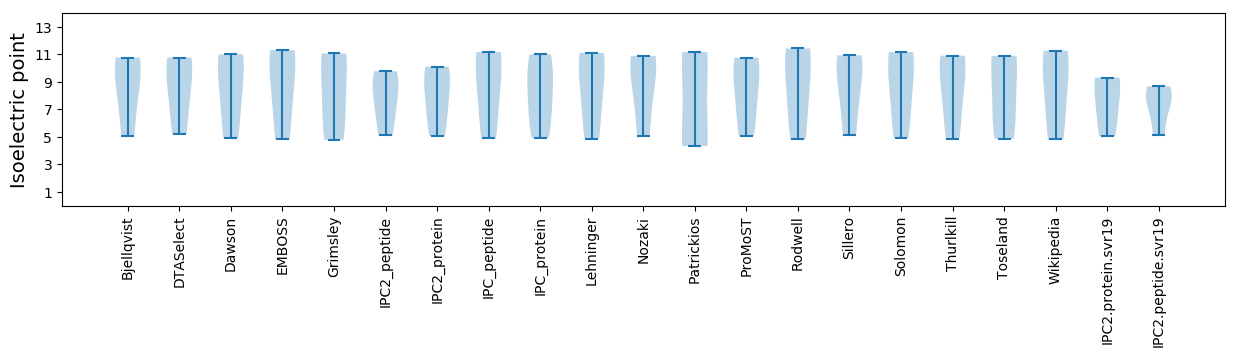
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4U8V8|A0A0B4U8V8_9VIRU Uncharacterized protein OS=Bromus-associated circular DNA virus 2 OX=1590155 PE=4 SV=1
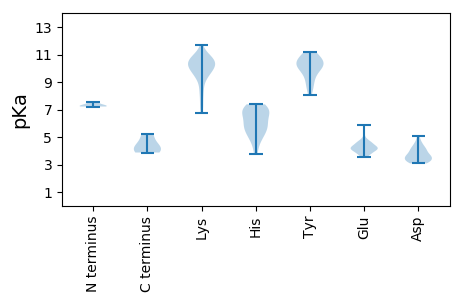
MM1 pKa = 7.29SQFKK5 pKa = 10.98SNSIWSEE12 pKa = 4.15VPDD15 pKa = 4.1HH16 pKa = 6.98EE17 pKa = 5.9KK18 pKa = 11.2DD19 pKa = 3.28LDD21 pKa = 4.09PPVSPLSLTTTNIEE35 pKa = 4.1EE36 pKa = 4.06HH37 pKa = 5.51NLRR40 pKa = 11.84YY41 pKa = 9.42RR42 pKa = 11.84RR43 pKa = 11.84CAGPVPPSRR52 pKa = 11.84ASTQSVASDD61 pKa = 3.52EE62 pKa = 4.56SRR64 pKa = 11.84ADD66 pKa = 3.54PSLVAVLSRR75 pKa = 11.84LDD77 pKa = 3.78LQDD80 pKa = 3.65AALDD84 pKa = 4.41EE85 pKa = 4.38ILARR89 pKa = 11.84LSLLHH94 pKa = 7.01RR95 pKa = 11.84EE96 pKa = 4.68FSRR99 pKa = 11.84PLPSPIISSTTQPEE113 pKa = 4.63FSATLPPGLLWW124 pKa = 4.36
MM1 pKa = 7.29SQFKK5 pKa = 10.98SNSIWSEE12 pKa = 4.15VPDD15 pKa = 4.1HH16 pKa = 6.98EE17 pKa = 5.9KK18 pKa = 11.2DD19 pKa = 3.28LDD21 pKa = 4.09PPVSPLSLTTTNIEE35 pKa = 4.1EE36 pKa = 4.06HH37 pKa = 5.51NLRR40 pKa = 11.84YY41 pKa = 9.42RR42 pKa = 11.84RR43 pKa = 11.84CAGPVPPSRR52 pKa = 11.84ASTQSVASDD61 pKa = 3.52EE62 pKa = 4.56SRR64 pKa = 11.84ADD66 pKa = 3.54PSLVAVLSRR75 pKa = 11.84LDD77 pKa = 3.78LQDD80 pKa = 3.65AALDD84 pKa = 4.41EE85 pKa = 4.38ILARR89 pKa = 11.84LSLLHH94 pKa = 7.01RR95 pKa = 11.84EE96 pKa = 4.68FSRR99 pKa = 11.84PLPSPIISSTTQPEE113 pKa = 4.63FSATLPPGLLWW124 pKa = 4.36
Molecular weight: 13.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4U8Q3|A0A0B4U8Q3_9VIRU Uncharacterized protein OS=Bromus-associated circular DNA virus 2 OX=1590155 PE=4 SV=1
MM1 pKa = 7.25VGGSRR6 pKa = 11.84SRR8 pKa = 11.84KK9 pKa = 8.53RR10 pKa = 11.84PRR12 pKa = 11.84SPGQSSVADD21 pKa = 3.89YY22 pKa = 10.26YY23 pKa = 10.88KK24 pKa = 10.49HH25 pKa = 6.91RR26 pKa = 11.84RR27 pKa = 11.84TQSPIPTMRR36 pKa = 11.84WARR39 pKa = 11.84AAFKK43 pKa = 10.58GKK45 pKa = 9.8YY46 pKa = 8.47AVRR49 pKa = 11.84RR50 pKa = 11.84KK51 pKa = 10.0RR52 pKa = 11.84RR53 pKa = 11.84VTRR56 pKa = 11.84RR57 pKa = 11.84SLSRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84LKK65 pKa = 10.26PSRR68 pKa = 11.84FTRR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84SRR75 pKa = 11.84RR76 pKa = 11.84NTRR79 pKa = 11.84PSKK82 pKa = 10.45SFASRR87 pKa = 11.84VLKK90 pKa = 9.61ATAVTNHH97 pKa = 7.38LIYY100 pKa = 9.88DD101 pKa = 3.75TAGILRR107 pKa = 11.84NVTTGVALVNVKK119 pKa = 10.46NPTLTQTDD127 pKa = 4.3FQAVQAKK134 pKa = 9.92LNNDD138 pKa = 3.33EE139 pKa = 4.67VLDD142 pKa = 4.06SVTNKK147 pKa = 10.11SFYY150 pKa = 10.27LRR152 pKa = 11.84HH153 pKa = 5.61ARR155 pKa = 11.84EE156 pKa = 4.07TYY158 pKa = 9.71TYY160 pKa = 10.28TNSSTNKK167 pKa = 7.46ITLKK171 pKa = 9.89IYY173 pKa = 9.96HH174 pKa = 5.88YY175 pKa = 10.4KK176 pKa = 10.78ARR178 pKa = 11.84FDD180 pKa = 4.37YY181 pKa = 11.0NISPQTLYY189 pKa = 11.18NQGFTDD195 pKa = 4.59DD196 pKa = 4.23GAIQATSVNSTPFMSNNFTTYY217 pKa = 10.56NKK219 pKa = 8.18ITRR222 pKa = 11.84VVTKK226 pKa = 9.97FLEE229 pKa = 4.51QGEE232 pKa = 4.64HH233 pKa = 3.8MTVYY237 pKa = 10.48QNQSSKK243 pKa = 10.78HH244 pKa = 5.38INSDD248 pKa = 3.27RR249 pKa = 11.84YY250 pKa = 10.46KK251 pKa = 10.8NSLVVAGSHH260 pKa = 4.56GHH262 pKa = 4.87VLFGVGTLGYY272 pKa = 8.76DD273 pKa = 3.54TVNTGITTGPVEE285 pKa = 4.53LMQRR289 pKa = 11.84LQRR292 pKa = 11.84HH293 pKa = 5.32ISYY296 pKa = 10.21SQLADD301 pKa = 3.2NTVRR305 pKa = 11.84NHH307 pKa = 6.7SDD309 pKa = 3.15DD310 pKa = 4.05ALATGLYY317 pKa = 10.82ANFRR321 pKa = 11.84FANSDD326 pKa = 3.1TGVAVTGNAGDD337 pKa = 4.05VIPP340 pKa = 5.25
MM1 pKa = 7.25VGGSRR6 pKa = 11.84SRR8 pKa = 11.84KK9 pKa = 8.53RR10 pKa = 11.84PRR12 pKa = 11.84SPGQSSVADD21 pKa = 3.89YY22 pKa = 10.26YY23 pKa = 10.88KK24 pKa = 10.49HH25 pKa = 6.91RR26 pKa = 11.84RR27 pKa = 11.84TQSPIPTMRR36 pKa = 11.84WARR39 pKa = 11.84AAFKK43 pKa = 10.58GKK45 pKa = 9.8YY46 pKa = 8.47AVRR49 pKa = 11.84RR50 pKa = 11.84KK51 pKa = 10.0RR52 pKa = 11.84RR53 pKa = 11.84VTRR56 pKa = 11.84RR57 pKa = 11.84SLSRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84LKK65 pKa = 10.26PSRR68 pKa = 11.84FTRR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84SRR75 pKa = 11.84RR76 pKa = 11.84NTRR79 pKa = 11.84PSKK82 pKa = 10.45SFASRR87 pKa = 11.84VLKK90 pKa = 9.61ATAVTNHH97 pKa = 7.38LIYY100 pKa = 9.88DD101 pKa = 3.75TAGILRR107 pKa = 11.84NVTTGVALVNVKK119 pKa = 10.46NPTLTQTDD127 pKa = 4.3FQAVQAKK134 pKa = 9.92LNNDD138 pKa = 3.33EE139 pKa = 4.67VLDD142 pKa = 4.06SVTNKK147 pKa = 10.11SFYY150 pKa = 10.27LRR152 pKa = 11.84HH153 pKa = 5.61ARR155 pKa = 11.84EE156 pKa = 4.07TYY158 pKa = 9.71TYY160 pKa = 10.28TNSSTNKK167 pKa = 7.46ITLKK171 pKa = 9.89IYY173 pKa = 9.96HH174 pKa = 5.88YY175 pKa = 10.4KK176 pKa = 10.78ARR178 pKa = 11.84FDD180 pKa = 4.37YY181 pKa = 11.0NISPQTLYY189 pKa = 11.18NQGFTDD195 pKa = 4.59DD196 pKa = 4.23GAIQATSVNSTPFMSNNFTTYY217 pKa = 10.56NKK219 pKa = 8.18ITRR222 pKa = 11.84VVTKK226 pKa = 9.97FLEE229 pKa = 4.51QGEE232 pKa = 4.64HH233 pKa = 3.8MTVYY237 pKa = 10.48QNQSSKK243 pKa = 10.78HH244 pKa = 5.38INSDD248 pKa = 3.27RR249 pKa = 11.84YY250 pKa = 10.46KK251 pKa = 10.8NSLVVAGSHH260 pKa = 4.56GHH262 pKa = 4.87VLFGVGTLGYY272 pKa = 8.76DD273 pKa = 3.54TVNTGITTGPVEE285 pKa = 4.53LMQRR289 pKa = 11.84LQRR292 pKa = 11.84HH293 pKa = 5.32ISYY296 pKa = 10.21SQLADD301 pKa = 3.2NTVRR305 pKa = 11.84NHH307 pKa = 6.7SDD309 pKa = 3.15DD310 pKa = 4.05ALATGLYY317 pKa = 10.82ANFRR321 pKa = 11.84FANSDD326 pKa = 3.1TGVAVTGNAGDD337 pKa = 4.05VIPP340 pKa = 5.25
Molecular weight: 38.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
907 |
124 |
340 |
226.8 |
25.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.056 ± 0.65 | 1.433 ± 0.804 |
4.851 ± 0.57 | 4.851 ± 1.526 |
3.087 ± 0.642 | 5.182 ± 0.612 |
2.646 ± 0.123 | 4.631 ± 0.569 |
6.615 ± 1.351 | 8.049 ± 1.545 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.544 ± 0.326 | 4.41 ± 1.236 |
5.513 ± 1.152 | 3.638 ± 0.199 |
8.049 ± 1.082 | 9.372 ± 1.339 |
7.718 ± 1.409 | 6.505 ± 0.62 |
1.654 ± 0.938 | 3.197 ± 0.896 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
