
Candidatus Enterovibrio luxaltus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Enterovibrio
Average proteome isoelectric point is 7.33
Get precalculated fractions of proteins
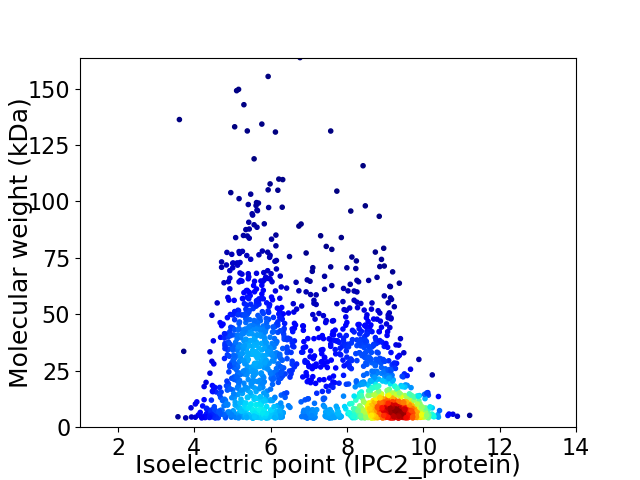
Virtual 2D-PAGE plot for 1986 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A291BC35|A0A291BC35_9GAMM Uncharacterized protein OS=Candidatus Enterovibrio luxaltus OX=1927128 GN=BTN50_2107 PE=4 SV=1
MM1 pKa = 7.26ATDD4 pKa = 4.59KK5 pKa = 9.33PTLTLIGDD13 pKa = 3.89SEE15 pKa = 4.61VIEE18 pKa = 5.22GEE20 pKa = 4.05DD21 pKa = 3.79ANYY24 pKa = 9.76TLAISEE30 pKa = 4.66APISDD35 pKa = 3.56LTVTVVVGHH44 pKa = 6.03KK45 pKa = 9.24TSEE48 pKa = 4.36DD49 pKa = 3.75DD50 pKa = 4.05DD51 pKa = 4.13VTPITHH57 pKa = 5.61QVVIAAGTTFTSFTVNTINDD77 pKa = 4.18GLNEE81 pKa = 4.21SVDD84 pKa = 3.38NDD86 pKa = 3.71VFTLSILGSSGGGFEE101 pKa = 4.76EE102 pKa = 5.54LPSTLTAIEE111 pKa = 3.99TVIRR115 pKa = 11.84DD116 pKa = 3.93DD117 pKa = 5.58AEE119 pKa = 4.46DD120 pKa = 3.65AAQLMEE126 pKa = 6.1DD127 pKa = 4.27IGTQSISGVLHH138 pKa = 6.01VLDD141 pKa = 4.81SNGLTPLEE149 pKa = 4.25FTNTTVVGNYY159 pKa = 10.17GSLTLVNGDD168 pKa = 3.28WTYY171 pKa = 11.98LLYY174 pKa = 8.92PTAQILNEE182 pKa = 4.23GEE184 pKa = 4.14NVTEE188 pKa = 5.29FIILTATNGTVQTIVISVIGTEE210 pKa = 4.21DD211 pKa = 3.52SAVVTGDD218 pKa = 3.83FIGSVDD224 pKa = 3.77VADD227 pKa = 4.94FSINTEE233 pKa = 3.38ASNNIRR239 pKa = 11.84ASNGEE244 pKa = 4.07IISNANSAWTTTSTMVNNVPYY265 pKa = 10.64VIATQWGSSGSTIISRR281 pKa = 11.84VNNDD285 pKa = 2.76GTLTEE290 pKa = 4.39TDD292 pKa = 3.86RR293 pKa = 11.84ITYY296 pKa = 7.62NQSSASVITTSGGNITADD314 pKa = 3.31VRR316 pKa = 11.84ALGILPEE323 pKa = 4.18SLGNGLTQSNAANIDD338 pKa = 3.55GQNTLFVTSQNSSSLTAWNISDD360 pKa = 4.84SGVLTINGGITNINAEE376 pKa = 4.63DD377 pKa = 3.8GQSYY381 pKa = 10.01IRR383 pKa = 11.84EE384 pKa = 4.25NITFEE389 pKa = 3.94GANGINYY396 pKa = 9.46IYY398 pKa = 9.29VTRR401 pKa = 11.84PGTDD405 pKa = 3.71GISKK409 pKa = 8.38FTYY412 pKa = 9.44SSKK415 pKa = 9.56TGEE418 pKa = 3.9MVNTNEE424 pKa = 4.04FVFGGEE430 pKa = 4.39TVSSIDD436 pKa = 3.21VCTTNDD442 pKa = 3.54YY443 pKa = 11.72NSFLAAASNDD453 pKa = 3.03AVRR456 pKa = 11.84IFAINQSTGALTLVEE471 pKa = 4.3AVVVPQGTSNSVNYY485 pKa = 9.84YY486 pKa = 7.92HH487 pKa = 7.09TNDD490 pKa = 3.1NRR492 pKa = 11.84SYY494 pKa = 10.76IIYY497 pKa = 10.7SNNTSNKK504 pKa = 9.63ASIFQMAEE512 pKa = 3.82NGSLTLTDD520 pKa = 4.37TIDD523 pKa = 3.65GAGHH527 pKa = 5.9YY528 pKa = 9.13FSSAGYY534 pKa = 9.39IDD536 pKa = 4.62GEE538 pKa = 4.25PVYY541 pKa = 10.87IMPNATAGVDD551 pKa = 3.78LYY553 pKa = 11.03TIGRR557 pKa = 11.84NGSFVLQTTVANISNDD573 pKa = 2.74WTPPVIVQTEE583 pKa = 3.95DD584 pKa = 2.71SSYY587 pKa = 11.32YY588 pKa = 10.79LVDD591 pKa = 3.85ADD593 pKa = 4.74GLASAIKK600 pKa = 9.25LTVGNTGDD608 pKa = 3.4GSVGNEE614 pKa = 3.77GVKK617 pKa = 10.72VSGSLHH623 pKa = 6.37ISDD626 pKa = 4.67IDD628 pKa = 4.01SEE630 pKa = 4.57DD631 pKa = 3.49FSSFEE636 pKa = 4.65NITITGTYY644 pKa = 7.03GTLVLVEE651 pKa = 4.17GTWIYY656 pKa = 10.29TLDD659 pKa = 3.73KK660 pKa = 10.83DD661 pKa = 3.85KK662 pKa = 11.46SSNILDD668 pKa = 3.79DD669 pKa = 5.76KK670 pKa = 10.28IAKK673 pKa = 9.73DD674 pKa = 3.86AFRR677 pKa = 11.84LTAIDD682 pKa = 4.32GTTQDD687 pKa = 4.0IIIDD691 pKa = 3.93VTGHH695 pKa = 4.61EE696 pKa = 4.3TALEE700 pKa = 4.19GGVYY704 pKa = 10.38GKK706 pKa = 8.47NTDD709 pKa = 3.73AIIVYY714 pKa = 8.9LSPQAASPGFIEE726 pKa = 4.94EE727 pKa = 4.21SSGDD731 pKa = 3.39VNFNLRR737 pKa = 11.84EE738 pKa = 4.0YY739 pKa = 10.95VADD742 pKa = 4.14LVDD745 pKa = 5.68DD746 pKa = 5.37ADD748 pKa = 4.67LSDD751 pKa = 3.64ARR753 pKa = 11.84KK754 pKa = 8.04TSMQIISLPNDD765 pKa = 3.21GVLYY769 pKa = 10.13YY770 pKa = 10.93YY771 pKa = 10.69NEE773 pKa = 4.61SGDD776 pKa = 3.77KK777 pKa = 10.56VPVVINNVLSDD788 pKa = 3.32EE789 pKa = 4.64TIFIFEE795 pKa = 4.81PNRR798 pKa = 11.84ASITFSQNDD807 pKa = 3.92IIIATQTTIISNGITVSGGIFSGTKK832 pKa = 9.58PDD834 pKa = 3.98DD835 pKa = 3.76SSTISPANIVWEE847 pKa = 4.53FQSGEE852 pKa = 4.06SGIGVGLDD860 pKa = 3.4RR861 pKa = 11.84EE862 pKa = 4.59ITSSLKK868 pKa = 10.36EE869 pKa = 3.72VLTIEE874 pKa = 4.51FTGSTNVTNADD885 pKa = 3.18IVVSSAFGNFSDD897 pKa = 4.22TRR899 pKa = 11.84SAQGKK904 pKa = 8.06GQLIAFRR911 pKa = 11.84DD912 pKa = 4.11GQVVGEE918 pKa = 3.77YY919 pKa = 10.33SYY921 pKa = 12.18ANLWEE926 pKa = 4.19ASGGDD931 pKa = 3.72GVATLNIVNSEE942 pKa = 4.05GFNEE946 pKa = 3.76LRR948 pKa = 11.84LYY950 pKa = 8.87ITANVNSNFSVTSISASGVNTTDD973 pKa = 2.89TQFTYY978 pKa = 10.84KK979 pKa = 10.62AINSFGNEE987 pKa = 3.9SEE989 pKa = 4.13IATVNLGEE997 pKa = 4.47NIIDD1001 pKa = 4.11SVTMPAISTVSGTLVIVNSTGVTAVFANTTKK1032 pKa = 10.51SGTYY1036 pKa = 10.57GSVTLMNGTWTYY1048 pKa = 9.52TLHH1051 pKa = 6.92PALLTVISNNQSIQDD1066 pKa = 4.36TITLTATDD1074 pKa = 3.79GTIAEE1079 pKa = 4.2VTIIISEE1086 pKa = 4.15SSNAIISYY1094 pKa = 7.62YY1095 pKa = 7.39TTIANNSLSGLIDD1108 pKa = 3.2MGSISFLGNNDD1119 pKa = 3.07TDD1121 pKa = 4.4NIYY1124 pKa = 11.03AGNNNDD1130 pKa = 5.32GILSLDD1136 pKa = 3.81GDD1138 pKa = 4.04DD1139 pKa = 4.02TVYY1142 pKa = 11.02SGAGDD1147 pKa = 4.6DD1148 pKa = 5.49IIDD1151 pKa = 4.33LGAGNDD1157 pKa = 3.91LVVGGAGNDD1166 pKa = 3.32ILIGGDD1172 pKa = 3.54GEE1174 pKa = 5.47DD1175 pKa = 3.84VFTFLNGNEE1184 pKa = 4.26GTNEE1188 pKa = 3.96VPTIDD1193 pKa = 5.28HH1194 pKa = 6.76IGDD1197 pKa = 3.79FNTNQDD1203 pKa = 3.78ALNVTDD1209 pKa = 5.22LLQGEE1214 pKa = 4.58TNDD1217 pKa = 5.28TIGQYY1222 pKa = 11.27LNIVMDD1228 pKa = 3.91SQGYY1232 pKa = 10.46AMLNISSMGNGMVDD1246 pKa = 3.24QQVVFDD1252 pKa = 4.27NLSVNDD1258 pKa = 3.98MATAYY1263 pKa = 10.58QIDD1266 pKa = 3.81ASGISADD1273 pKa = 4.17QISTLIIDD1281 pKa = 3.84AMIVQSRR1288 pKa = 11.84IIIDD1292 pKa = 3.33
MM1 pKa = 7.26ATDD4 pKa = 4.59KK5 pKa = 9.33PTLTLIGDD13 pKa = 3.89SEE15 pKa = 4.61VIEE18 pKa = 5.22GEE20 pKa = 4.05DD21 pKa = 3.79ANYY24 pKa = 9.76TLAISEE30 pKa = 4.66APISDD35 pKa = 3.56LTVTVVVGHH44 pKa = 6.03KK45 pKa = 9.24TSEE48 pKa = 4.36DD49 pKa = 3.75DD50 pKa = 4.05DD51 pKa = 4.13VTPITHH57 pKa = 5.61QVVIAAGTTFTSFTVNTINDD77 pKa = 4.18GLNEE81 pKa = 4.21SVDD84 pKa = 3.38NDD86 pKa = 3.71VFTLSILGSSGGGFEE101 pKa = 4.76EE102 pKa = 5.54LPSTLTAIEE111 pKa = 3.99TVIRR115 pKa = 11.84DD116 pKa = 3.93DD117 pKa = 5.58AEE119 pKa = 4.46DD120 pKa = 3.65AAQLMEE126 pKa = 6.1DD127 pKa = 4.27IGTQSISGVLHH138 pKa = 6.01VLDD141 pKa = 4.81SNGLTPLEE149 pKa = 4.25FTNTTVVGNYY159 pKa = 10.17GSLTLVNGDD168 pKa = 3.28WTYY171 pKa = 11.98LLYY174 pKa = 8.92PTAQILNEE182 pKa = 4.23GEE184 pKa = 4.14NVTEE188 pKa = 5.29FIILTATNGTVQTIVISVIGTEE210 pKa = 4.21DD211 pKa = 3.52SAVVTGDD218 pKa = 3.83FIGSVDD224 pKa = 3.77VADD227 pKa = 4.94FSINTEE233 pKa = 3.38ASNNIRR239 pKa = 11.84ASNGEE244 pKa = 4.07IISNANSAWTTTSTMVNNVPYY265 pKa = 10.64VIATQWGSSGSTIISRR281 pKa = 11.84VNNDD285 pKa = 2.76GTLTEE290 pKa = 4.39TDD292 pKa = 3.86RR293 pKa = 11.84ITYY296 pKa = 7.62NQSSASVITTSGGNITADD314 pKa = 3.31VRR316 pKa = 11.84ALGILPEE323 pKa = 4.18SLGNGLTQSNAANIDD338 pKa = 3.55GQNTLFVTSQNSSSLTAWNISDD360 pKa = 4.84SGVLTINGGITNINAEE376 pKa = 4.63DD377 pKa = 3.8GQSYY381 pKa = 10.01IRR383 pKa = 11.84EE384 pKa = 4.25NITFEE389 pKa = 3.94GANGINYY396 pKa = 9.46IYY398 pKa = 9.29VTRR401 pKa = 11.84PGTDD405 pKa = 3.71GISKK409 pKa = 8.38FTYY412 pKa = 9.44SSKK415 pKa = 9.56TGEE418 pKa = 3.9MVNTNEE424 pKa = 4.04FVFGGEE430 pKa = 4.39TVSSIDD436 pKa = 3.21VCTTNDD442 pKa = 3.54YY443 pKa = 11.72NSFLAAASNDD453 pKa = 3.03AVRR456 pKa = 11.84IFAINQSTGALTLVEE471 pKa = 4.3AVVVPQGTSNSVNYY485 pKa = 9.84YY486 pKa = 7.92HH487 pKa = 7.09TNDD490 pKa = 3.1NRR492 pKa = 11.84SYY494 pKa = 10.76IIYY497 pKa = 10.7SNNTSNKK504 pKa = 9.63ASIFQMAEE512 pKa = 3.82NGSLTLTDD520 pKa = 4.37TIDD523 pKa = 3.65GAGHH527 pKa = 5.9YY528 pKa = 9.13FSSAGYY534 pKa = 9.39IDD536 pKa = 4.62GEE538 pKa = 4.25PVYY541 pKa = 10.87IMPNATAGVDD551 pKa = 3.78LYY553 pKa = 11.03TIGRR557 pKa = 11.84NGSFVLQTTVANISNDD573 pKa = 2.74WTPPVIVQTEE583 pKa = 3.95DD584 pKa = 2.71SSYY587 pKa = 11.32YY588 pKa = 10.79LVDD591 pKa = 3.85ADD593 pKa = 4.74GLASAIKK600 pKa = 9.25LTVGNTGDD608 pKa = 3.4GSVGNEE614 pKa = 3.77GVKK617 pKa = 10.72VSGSLHH623 pKa = 6.37ISDD626 pKa = 4.67IDD628 pKa = 4.01SEE630 pKa = 4.57DD631 pKa = 3.49FSSFEE636 pKa = 4.65NITITGTYY644 pKa = 7.03GTLVLVEE651 pKa = 4.17GTWIYY656 pKa = 10.29TLDD659 pKa = 3.73KK660 pKa = 10.83DD661 pKa = 3.85KK662 pKa = 11.46SSNILDD668 pKa = 3.79DD669 pKa = 5.76KK670 pKa = 10.28IAKK673 pKa = 9.73DD674 pKa = 3.86AFRR677 pKa = 11.84LTAIDD682 pKa = 4.32GTTQDD687 pKa = 4.0IIIDD691 pKa = 3.93VTGHH695 pKa = 4.61EE696 pKa = 4.3TALEE700 pKa = 4.19GGVYY704 pKa = 10.38GKK706 pKa = 8.47NTDD709 pKa = 3.73AIIVYY714 pKa = 8.9LSPQAASPGFIEE726 pKa = 4.94EE727 pKa = 4.21SSGDD731 pKa = 3.39VNFNLRR737 pKa = 11.84EE738 pKa = 4.0YY739 pKa = 10.95VADD742 pKa = 4.14LVDD745 pKa = 5.68DD746 pKa = 5.37ADD748 pKa = 4.67LSDD751 pKa = 3.64ARR753 pKa = 11.84KK754 pKa = 8.04TSMQIISLPNDD765 pKa = 3.21GVLYY769 pKa = 10.13YY770 pKa = 10.93YY771 pKa = 10.69NEE773 pKa = 4.61SGDD776 pKa = 3.77KK777 pKa = 10.56VPVVINNVLSDD788 pKa = 3.32EE789 pKa = 4.64TIFIFEE795 pKa = 4.81PNRR798 pKa = 11.84ASITFSQNDD807 pKa = 3.92IIIATQTTIISNGITVSGGIFSGTKK832 pKa = 9.58PDD834 pKa = 3.98DD835 pKa = 3.76SSTISPANIVWEE847 pKa = 4.53FQSGEE852 pKa = 4.06SGIGVGLDD860 pKa = 3.4RR861 pKa = 11.84EE862 pKa = 4.59ITSSLKK868 pKa = 10.36EE869 pKa = 3.72VLTIEE874 pKa = 4.51FTGSTNVTNADD885 pKa = 3.18IVVSSAFGNFSDD897 pKa = 4.22TRR899 pKa = 11.84SAQGKK904 pKa = 8.06GQLIAFRR911 pKa = 11.84DD912 pKa = 4.11GQVVGEE918 pKa = 3.77YY919 pKa = 10.33SYY921 pKa = 12.18ANLWEE926 pKa = 4.19ASGGDD931 pKa = 3.72GVATLNIVNSEE942 pKa = 4.05GFNEE946 pKa = 3.76LRR948 pKa = 11.84LYY950 pKa = 8.87ITANVNSNFSVTSISASGVNTTDD973 pKa = 2.89TQFTYY978 pKa = 10.84KK979 pKa = 10.62AINSFGNEE987 pKa = 3.9SEE989 pKa = 4.13IATVNLGEE997 pKa = 4.47NIIDD1001 pKa = 4.11SVTMPAISTVSGTLVIVNSTGVTAVFANTTKK1032 pKa = 10.51SGTYY1036 pKa = 10.57GSVTLMNGTWTYY1048 pKa = 9.52TLHH1051 pKa = 6.92PALLTVISNNQSIQDD1066 pKa = 4.36TITLTATDD1074 pKa = 3.79GTIAEE1079 pKa = 4.2VTIIISEE1086 pKa = 4.15SSNAIISYY1094 pKa = 7.62YY1095 pKa = 7.39TTIANNSLSGLIDD1108 pKa = 3.2MGSISFLGNNDD1119 pKa = 3.07TDD1121 pKa = 4.4NIYY1124 pKa = 11.03AGNNNDD1130 pKa = 5.32GILSLDD1136 pKa = 3.81GDD1138 pKa = 4.04DD1139 pKa = 4.02TVYY1142 pKa = 11.02SGAGDD1147 pKa = 4.6DD1148 pKa = 5.49IIDD1151 pKa = 4.33LGAGNDD1157 pKa = 3.91LVVGGAGNDD1166 pKa = 3.32ILIGGDD1172 pKa = 3.54GEE1174 pKa = 5.47DD1175 pKa = 3.84VFTFLNGNEE1184 pKa = 4.26GTNEE1188 pKa = 3.96VPTIDD1193 pKa = 5.28HH1194 pKa = 6.76IGDD1197 pKa = 3.79FNTNQDD1203 pKa = 3.78ALNVTDD1209 pKa = 5.22LLQGEE1214 pKa = 4.58TNDD1217 pKa = 5.28TIGQYY1222 pKa = 11.27LNIVMDD1228 pKa = 3.91SQGYY1232 pKa = 10.46AMLNISSMGNGMVDD1246 pKa = 3.24QQVVFDD1252 pKa = 4.27NLSVNDD1258 pKa = 3.98MATAYY1263 pKa = 10.58QIDD1266 pKa = 3.81ASGISADD1273 pKa = 4.17QISTLIIDD1281 pKa = 3.84AMIVQSRR1288 pKa = 11.84IIIDD1292 pKa = 3.33
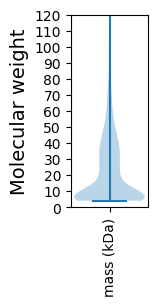
Molecular weight: 136.25 kDa
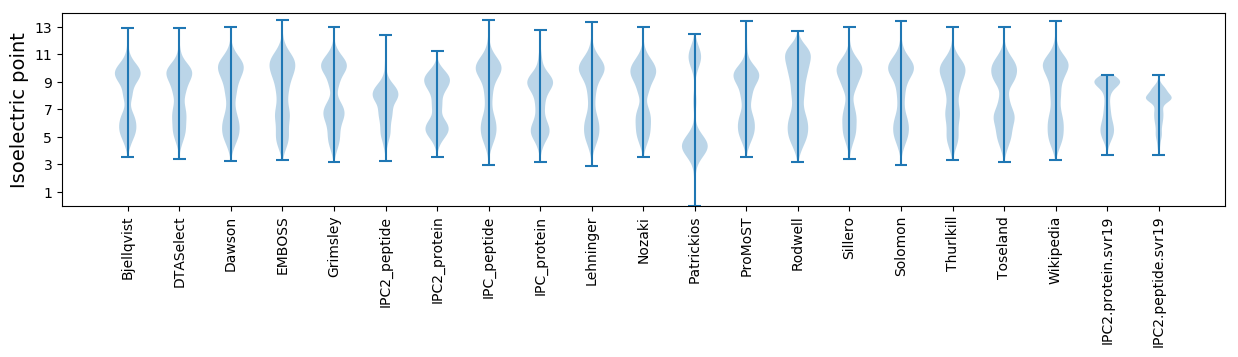
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A291B958|A0A291B958_9GAMM DNA gyrase subunit B OS=Candidatus Enterovibrio luxaltus OX=1927128 GN=gyrB PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.12RR14 pKa = 11.84SHH16 pKa = 5.87GFRR19 pKa = 11.84VRR21 pKa = 11.84MATKK25 pKa = 10.06HH26 pKa = 4.61GRR28 pKa = 11.84KK29 pKa = 9.31IIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.91GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSKK44 pKa = 10.84
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.12RR14 pKa = 11.84SHH16 pKa = 5.87GFRR19 pKa = 11.84VRR21 pKa = 11.84MATKK25 pKa = 10.06HH26 pKa = 4.61GRR28 pKa = 11.84KK29 pKa = 9.31IIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.91GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSKK44 pKa = 10.84
Molecular weight: 5.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
410369 |
37 |
1488 |
206.6 |
23.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.689 ± 0.065 | 1.249 ± 0.024 |
4.955 ± 0.047 | 5.651 ± 0.059 |
4.099 ± 0.042 | 6.459 ± 0.065 |
2.417 ± 0.034 | 7.79 ± 0.052 |
6.11 ± 0.063 | 10.215 ± 0.074 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.049 ± 0.036 | 5.018 ± 0.046 |
3.511 ± 0.034 | 3.996 ± 0.043 |
4.901 ± 0.057 | 6.369 ± 0.054 |
5.605 ± 0.045 | 6.855 ± 0.049 |
1.063 ± 0.023 | 2.999 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
