
Amphiprion ocellaris (Clown anemonefish)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Neoteleostei; Eurypterygia; Ctenosquamata; Acanthomorphata;
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 31745 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
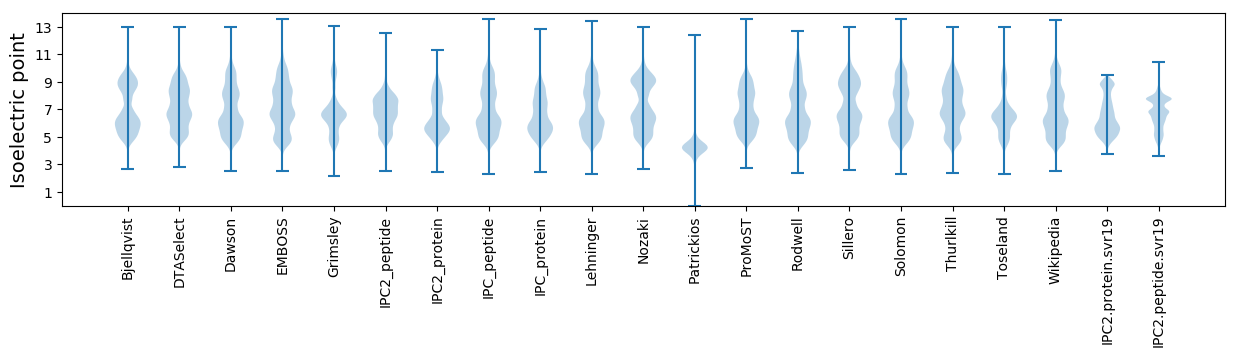
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A3Q1CDX7|A0A3Q1CDX7_AMPOC Hydroxycarboxylic acid receptor 1-3 OS=Amphiprion ocellaris OX=80972 PE=3 SV=1
MM1 pKa = 7.66LCTPSILDD9 pKa = 3.94HH10 pKa = 7.09PPSPMYY16 pKa = 9.87TFSVTAEE23 pKa = 4.12EE24 pKa = 4.22EE25 pKa = 4.04QGLDD29 pKa = 3.69GMDD32 pKa = 3.03WLDD35 pKa = 3.58FTSGRR40 pKa = 11.84EE41 pKa = 3.94KK42 pKa = 11.09DD43 pKa = 3.85EE44 pKa = 4.53EE45 pKa = 4.55PPTLAPLAPQTPSSVFSTDD64 pKa = 3.55FLDD67 pKa = 6.07GYY69 pKa = 10.27DD70 pKa = 3.65LHH72 pKa = 8.36IHH74 pKa = 6.97LDD76 pKa = 3.51SCLL79 pKa = 3.42
MM1 pKa = 7.66LCTPSILDD9 pKa = 3.94HH10 pKa = 7.09PPSPMYY16 pKa = 9.87TFSVTAEE23 pKa = 4.12EE24 pKa = 4.22EE25 pKa = 4.04QGLDD29 pKa = 3.69GMDD32 pKa = 3.03WLDD35 pKa = 3.58FTSGRR40 pKa = 11.84EE41 pKa = 3.94KK42 pKa = 11.09DD43 pKa = 3.85EE44 pKa = 4.53EE45 pKa = 4.55PPTLAPLAPQTPSSVFSTDD64 pKa = 3.55FLDD67 pKa = 6.07GYY69 pKa = 10.27DD70 pKa = 3.65LHH72 pKa = 8.36IHH74 pKa = 6.97LDD76 pKa = 3.51SCLL79 pKa = 3.42
Molecular weight: 8.76 kDa
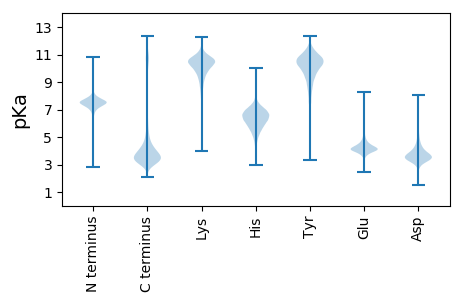
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q1B719|A0A3Q1B719_AMPOC Ribosomal protein L7 OS=Amphiprion ocellaris OX=80972 PE=3 SV=1
MM1 pKa = 6.59EE2 pKa = 4.24QFYY5 pKa = 10.8IEE7 pKa = 4.4CTRR10 pKa = 11.84GSSTPPRR17 pKa = 11.84GSSTPSRR24 pKa = 11.84GSSAPPRR31 pKa = 11.84GSSAPSRR38 pKa = 11.84QSSTPPRR45 pKa = 11.84QSSTPPRR52 pKa = 11.84GSSTPPRR59 pKa = 11.84GSSTPSRR66 pKa = 11.84GSSTPPRR73 pKa = 11.84GSSTPSRR80 pKa = 11.84GSSTPPRR87 pKa = 11.84GSSTPPRR94 pKa = 11.84GSSTPPRR101 pKa = 11.84GSSTPPRR108 pKa = 11.84GSSTPPRR115 pKa = 11.84GSSTPSRR122 pKa = 11.84QSSTPPRR129 pKa = 11.84GSSTPSRR136 pKa = 11.84QSSTPPRR143 pKa = 11.84GSSTPSRR150 pKa = 11.84QSSTPPRR157 pKa = 11.84GSSTPPRR164 pKa = 11.84GSSTPSRR171 pKa = 11.84QSSTPPRR178 pKa = 11.84GSSTPPRR185 pKa = 11.84RR186 pKa = 11.84SSTPPRR192 pKa = 11.84GSSTPPRR199 pKa = 11.84GSSAPSRR206 pKa = 11.84QSSTPPRR213 pKa = 11.84GSSAPTRR220 pKa = 11.84GSSTPSRR227 pKa = 11.84GSSTPPRR234 pKa = 11.84GSSSPPRR241 pKa = 11.84GSSTPSRR248 pKa = 11.84GSSTPPRR255 pKa = 11.84GSSTPSRR262 pKa = 11.84GSSTPPRR269 pKa = 11.84GSSTPPRR276 pKa = 11.84GSSTPPRR283 pKa = 11.84GSSTPPRR290 pKa = 11.84GSSTPPRR297 pKa = 11.84GSSTPSRR304 pKa = 11.84QSSTPPRR311 pKa = 11.84GSSTPSRR318 pKa = 11.84QSSTPPRR325 pKa = 11.84GSSTPSRR332 pKa = 11.84QSSTPPRR339 pKa = 11.84GSSTPPRR346 pKa = 11.84GSSTPSRR353 pKa = 11.84QSSTPPRR360 pKa = 11.84GSSTPPRR367 pKa = 11.84RR368 pKa = 11.84SSTPPRR374 pKa = 11.84GSSTPPRR381 pKa = 11.84GSSAPSRR388 pKa = 11.84QSSTPPRR395 pKa = 11.84GSSTPPRR402 pKa = 4.56
MM1 pKa = 6.59EE2 pKa = 4.24QFYY5 pKa = 10.8IEE7 pKa = 4.4CTRR10 pKa = 11.84GSSTPPRR17 pKa = 11.84GSSTPSRR24 pKa = 11.84GSSAPPRR31 pKa = 11.84GSSAPSRR38 pKa = 11.84QSSTPPRR45 pKa = 11.84QSSTPPRR52 pKa = 11.84GSSTPPRR59 pKa = 11.84GSSTPSRR66 pKa = 11.84GSSTPPRR73 pKa = 11.84GSSTPSRR80 pKa = 11.84GSSTPPRR87 pKa = 11.84GSSTPPRR94 pKa = 11.84GSSTPPRR101 pKa = 11.84GSSTPPRR108 pKa = 11.84GSSTPPRR115 pKa = 11.84GSSTPSRR122 pKa = 11.84QSSTPPRR129 pKa = 11.84GSSTPSRR136 pKa = 11.84QSSTPPRR143 pKa = 11.84GSSTPSRR150 pKa = 11.84QSSTPPRR157 pKa = 11.84GSSTPPRR164 pKa = 11.84GSSTPSRR171 pKa = 11.84QSSTPPRR178 pKa = 11.84GSSTPPRR185 pKa = 11.84RR186 pKa = 11.84SSTPPRR192 pKa = 11.84GSSTPPRR199 pKa = 11.84GSSAPSRR206 pKa = 11.84QSSTPPRR213 pKa = 11.84GSSAPTRR220 pKa = 11.84GSSTPSRR227 pKa = 11.84GSSTPPRR234 pKa = 11.84GSSSPPRR241 pKa = 11.84GSSTPSRR248 pKa = 11.84GSSTPPRR255 pKa = 11.84GSSTPSRR262 pKa = 11.84GSSTPPRR269 pKa = 11.84GSSTPPRR276 pKa = 11.84GSSTPPRR283 pKa = 11.84GSSTPPRR290 pKa = 11.84GSSTPPRR297 pKa = 11.84GSSTPSRR304 pKa = 11.84QSSTPPRR311 pKa = 11.84GSSTPSRR318 pKa = 11.84QSSTPPRR325 pKa = 11.84GSSTPSRR332 pKa = 11.84QSSTPPRR339 pKa = 11.84GSSTPPRR346 pKa = 11.84GSSTPSRR353 pKa = 11.84QSSTPPRR360 pKa = 11.84GSSTPPRR367 pKa = 11.84RR368 pKa = 11.84SSTPPRR374 pKa = 11.84GSSTPPRR381 pKa = 11.84GSSAPSRR388 pKa = 11.84QSSTPPRR395 pKa = 11.84GSSTPPRR402 pKa = 4.56
Molecular weight: 40.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
17832489 |
17 |
7970 |
561.7 |
62.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.525 ± 0.011 | 2.247 ± 0.011 |
5.234 ± 0.01 | 6.826 ± 0.018 |
3.66 ± 0.011 | 6.29 ± 0.013 |
2.655 ± 0.008 | 4.405 ± 0.011 |
5.601 ± 0.015 | 9.524 ± 0.017 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.392 ± 0.006 | 3.831 ± 0.007 |
5.697 ± 0.02 | 4.769 ± 0.012 |
5.625 ± 0.012 | 8.755 ± 0.02 |
5.658 ± 0.011 | 6.386 ± 0.013 |
1.147 ± 0.005 | 2.773 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
