
Capybara microvirus Cap3_SP_433
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.63
Get precalculated fractions of proteins
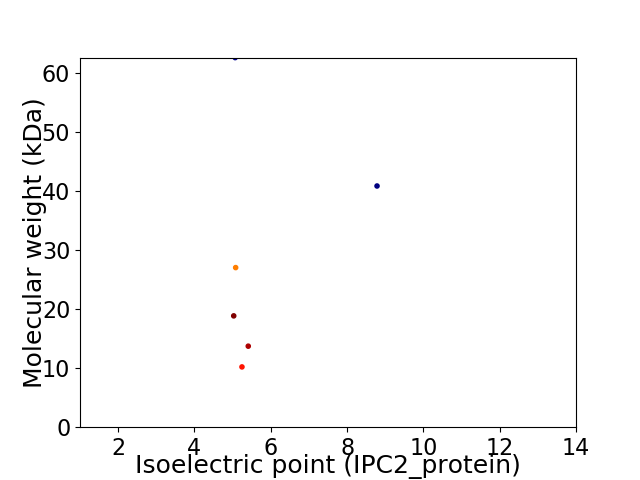
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W633|A0A4P8W633_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_433 OX=2585451 PE=4 SV=1
MM1 pKa = 7.46AAASTAAGIGSILSGSGSMLSGSVSNGSSYY31 pKa = 10.02STSSDD36 pKa = 2.48RR37 pKa = 11.84GYY39 pKa = 11.03SDD41 pKa = 3.93SSSSSWEE48 pKa = 3.93GGSSYY53 pKa = 11.26GSADD57 pKa = 3.51SYY59 pKa = 11.83SSANSASAYY68 pKa = 9.71YY69 pKa = 10.67DD70 pKa = 3.42SAQSEE75 pKa = 4.15NWANSWTEE83 pKa = 3.36ASAANEE89 pKa = 3.92QAAKK93 pKa = 9.53SAYY96 pKa = 9.84YY97 pKa = 10.57SNQLQEE103 pKa = 3.77RR104 pKa = 11.84WYY106 pKa = 10.39KK107 pKa = 10.87EE108 pKa = 3.48QMQYY112 pKa = 11.04NAEE115 pKa = 3.97QAKK118 pKa = 6.4TQRR121 pKa = 11.84EE122 pKa = 4.28YY123 pKa = 11.22EE124 pKa = 3.86EE125 pKa = 5.49RR126 pKa = 11.84MANTAYY132 pKa = 10.0QRR134 pKa = 11.84AVADD138 pKa = 4.63LKK140 pKa = 11.3AAGLNPILAAMLGGSATPAASAASVMAPSTSKK172 pKa = 11.31AEE174 pKa = 4.18TYY176 pKa = 9.7MNSGSSAYY184 pKa = 10.17GYY186 pKa = 9.86GKK188 pKa = 10.4SYY190 pKa = 11.42GEE192 pKa = 4.19GWSNSNQYY200 pKa = 10.44SEE202 pKa = 4.52SHH204 pKa = 6.16EE205 pKa = 4.23KK206 pKa = 10.62SGSSWGGGSQSTSHH220 pKa = 6.04SQNDD224 pKa = 3.53AKK226 pKa = 10.9SVSRR230 pKa = 11.84SSSTASSNLVNGLSAIGAGIKK251 pKa = 8.82EE252 pKa = 4.01LWYY255 pKa = 10.69KK256 pKa = 10.94GEE258 pKa = 3.97EE259 pKa = 4.02RR260 pKa = 4.48
MM1 pKa = 7.46AAASTAAGIGSILSGSGSMLSGSVSNGSSYY31 pKa = 10.02STSSDD36 pKa = 2.48RR37 pKa = 11.84GYY39 pKa = 11.03SDD41 pKa = 3.93SSSSSWEE48 pKa = 3.93GGSSYY53 pKa = 11.26GSADD57 pKa = 3.51SYY59 pKa = 11.83SSANSASAYY68 pKa = 9.71YY69 pKa = 10.67DD70 pKa = 3.42SAQSEE75 pKa = 4.15NWANSWTEE83 pKa = 3.36ASAANEE89 pKa = 3.92QAAKK93 pKa = 9.53SAYY96 pKa = 9.84YY97 pKa = 10.57SNQLQEE103 pKa = 3.77RR104 pKa = 11.84WYY106 pKa = 10.39KK107 pKa = 10.87EE108 pKa = 3.48QMQYY112 pKa = 11.04NAEE115 pKa = 3.97QAKK118 pKa = 6.4TQRR121 pKa = 11.84EE122 pKa = 4.28YY123 pKa = 11.22EE124 pKa = 3.86EE125 pKa = 5.49RR126 pKa = 11.84MANTAYY132 pKa = 10.0QRR134 pKa = 11.84AVADD138 pKa = 4.63LKK140 pKa = 11.3AAGLNPILAAMLGGSATPAASAASVMAPSTSKK172 pKa = 11.31AEE174 pKa = 4.18TYY176 pKa = 9.7MNSGSSAYY184 pKa = 10.17GYY186 pKa = 9.86GKK188 pKa = 10.4SYY190 pKa = 11.42GEE192 pKa = 4.19GWSNSNQYY200 pKa = 10.44SEE202 pKa = 4.52SHH204 pKa = 6.16EE205 pKa = 4.23KK206 pKa = 10.62SGSSWGGGSQSTSHH220 pKa = 6.04SQNDD224 pKa = 3.53AKK226 pKa = 10.9SVSRR230 pKa = 11.84SSSTASSNLVNGLSAIGAGIKK251 pKa = 8.82EE252 pKa = 4.01LWYY255 pKa = 10.69KK256 pKa = 10.94GEE258 pKa = 3.97EE259 pKa = 4.02RR260 pKa = 4.48
Molecular weight: 27.02 kDa
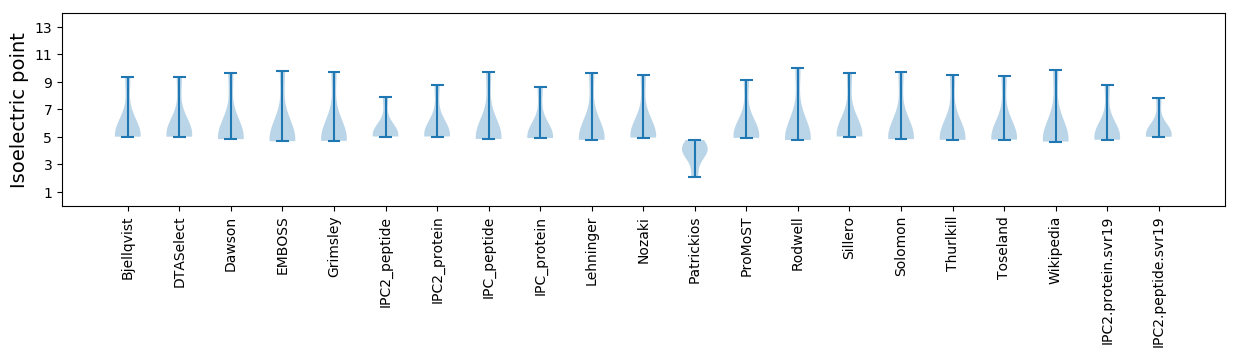
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W4Z4|A0A4P8W4Z4_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_433 OX=2585451 PE=3 SV=1
MM1 pKa = 7.34SCYY4 pKa = 9.61HH5 pKa = 6.55PKK7 pKa = 10.22IIIITEE13 pKa = 3.81EE14 pKa = 3.63AKK16 pKa = 10.79LNKK19 pKa = 9.6NGKK22 pKa = 9.2IGSLVKK28 pKa = 10.29FVEE31 pKa = 5.01DD32 pKa = 3.51EE33 pKa = 4.39KK34 pKa = 11.64AWGGYY39 pKa = 7.47EE40 pKa = 4.36NIEE43 pKa = 4.19KK44 pKa = 10.48FNAKK48 pKa = 10.26AEE50 pKa = 4.22EE51 pKa = 3.91QRR53 pKa = 11.84KK54 pKa = 9.86KK55 pKa = 11.02YY56 pKa = 10.31GLKK59 pKa = 8.81KK60 pKa = 9.07TYY62 pKa = 9.79ATMAPCGKK70 pKa = 10.01CQGCRR75 pKa = 11.84FKK77 pKa = 10.84QSKK80 pKa = 8.2EE81 pKa = 3.41WAVRR85 pKa = 11.84IEE87 pKa = 4.48KK88 pKa = 9.66EE89 pKa = 3.8AKK91 pKa = 9.31QYY93 pKa = 8.79KK94 pKa = 9.55HH95 pKa = 6.47NYY97 pKa = 8.74FVTLTYY103 pKa = 10.83NDD105 pKa = 3.43EE106 pKa = 4.12NLPIKK111 pKa = 10.31EE112 pKa = 4.9GYY114 pKa = 9.12PEE116 pKa = 4.36SKK118 pKa = 9.36GTLKK122 pKa = 10.64KK123 pKa = 10.24EE124 pKa = 3.67HH125 pKa = 7.01TIQFMKK131 pKa = 10.52SIRR134 pKa = 11.84QSIKK138 pKa = 9.1RR139 pKa = 11.84HH140 pKa = 5.08YY141 pKa = 9.37NHH143 pKa = 7.49DD144 pKa = 3.98GIKK147 pKa = 10.24FYY149 pKa = 11.32LCGEE153 pKa = 4.25YY154 pKa = 10.47GSKK157 pKa = 10.83GEE159 pKa = 4.11RR160 pKa = 11.84PHH162 pKa = 6.65YY163 pKa = 9.65HH164 pKa = 7.24IIMMNMPEE172 pKa = 4.15LEE174 pKa = 4.13KK175 pKa = 11.27NKK177 pKa = 10.45LVGQNTKK184 pKa = 9.61TKK186 pKa = 10.8DD187 pKa = 3.27PYY189 pKa = 9.69FTNDD193 pKa = 3.67LFDD196 pKa = 4.22SAWSKK201 pKa = 11.22GFVIIGKK208 pKa = 7.96VNWNTISYY216 pKa = 8.94VAGYY220 pKa = 8.07CQKK223 pKa = 11.14KK224 pKa = 10.28LFGEE228 pKa = 4.3VGKK231 pKa = 10.62KK232 pKa = 9.15IYY234 pKa = 9.81EE235 pKa = 4.2MKK237 pKa = 10.39GQEE240 pKa = 4.2PIYY243 pKa = 10.27ATMSRR248 pKa = 11.84NPGIGRR254 pKa = 11.84NYY256 pKa = 9.86YY257 pKa = 9.88EE258 pKa = 4.36EE259 pKa = 4.21NKK261 pKa = 8.35EE262 pKa = 3.96TIYY265 pKa = 10.69RR266 pKa = 11.84YY267 pKa = 10.74DD268 pKa = 3.38EE269 pKa = 4.56VITSKK274 pKa = 10.76GKK276 pKa = 9.06SVKK279 pKa = 9.79PPSYY283 pKa = 10.24FDD285 pKa = 3.53RR286 pKa = 11.84LMDD289 pKa = 3.91IGEE292 pKa = 4.31EE293 pKa = 3.81DD294 pKa = 4.12SIILKK299 pKa = 8.94EE300 pKa = 4.09LKK302 pKa = 9.73EE303 pKa = 3.92KK304 pKa = 10.69RR305 pKa = 11.84RR306 pKa = 11.84EE307 pKa = 3.92KK308 pKa = 11.06SNNALKK314 pKa = 10.36QKK316 pKa = 10.09YY317 pKa = 9.17AQTTLTIKK325 pKa = 10.3EE326 pKa = 3.86QLALEE331 pKa = 4.24EE332 pKa = 4.05RR333 pKa = 11.84TAVEE337 pKa = 4.21KK338 pKa = 10.73LKK340 pKa = 10.68KK341 pKa = 9.98YY342 pKa = 10.26NRR344 pKa = 11.84DD345 pKa = 3.55RR346 pKa = 11.84IGQQ349 pKa = 3.55
MM1 pKa = 7.34SCYY4 pKa = 9.61HH5 pKa = 6.55PKK7 pKa = 10.22IIIITEE13 pKa = 3.81EE14 pKa = 3.63AKK16 pKa = 10.79LNKK19 pKa = 9.6NGKK22 pKa = 9.2IGSLVKK28 pKa = 10.29FVEE31 pKa = 5.01DD32 pKa = 3.51EE33 pKa = 4.39KK34 pKa = 11.64AWGGYY39 pKa = 7.47EE40 pKa = 4.36NIEE43 pKa = 4.19KK44 pKa = 10.48FNAKK48 pKa = 10.26AEE50 pKa = 4.22EE51 pKa = 3.91QRR53 pKa = 11.84KK54 pKa = 9.86KK55 pKa = 11.02YY56 pKa = 10.31GLKK59 pKa = 8.81KK60 pKa = 9.07TYY62 pKa = 9.79ATMAPCGKK70 pKa = 10.01CQGCRR75 pKa = 11.84FKK77 pKa = 10.84QSKK80 pKa = 8.2EE81 pKa = 3.41WAVRR85 pKa = 11.84IEE87 pKa = 4.48KK88 pKa = 9.66EE89 pKa = 3.8AKK91 pKa = 9.31QYY93 pKa = 8.79KK94 pKa = 9.55HH95 pKa = 6.47NYY97 pKa = 8.74FVTLTYY103 pKa = 10.83NDD105 pKa = 3.43EE106 pKa = 4.12NLPIKK111 pKa = 10.31EE112 pKa = 4.9GYY114 pKa = 9.12PEE116 pKa = 4.36SKK118 pKa = 9.36GTLKK122 pKa = 10.64KK123 pKa = 10.24EE124 pKa = 3.67HH125 pKa = 7.01TIQFMKK131 pKa = 10.52SIRR134 pKa = 11.84QSIKK138 pKa = 9.1RR139 pKa = 11.84HH140 pKa = 5.08YY141 pKa = 9.37NHH143 pKa = 7.49DD144 pKa = 3.98GIKK147 pKa = 10.24FYY149 pKa = 11.32LCGEE153 pKa = 4.25YY154 pKa = 10.47GSKK157 pKa = 10.83GEE159 pKa = 4.11RR160 pKa = 11.84PHH162 pKa = 6.65YY163 pKa = 9.65HH164 pKa = 7.24IIMMNMPEE172 pKa = 4.15LEE174 pKa = 4.13KK175 pKa = 11.27NKK177 pKa = 10.45LVGQNTKK184 pKa = 9.61TKK186 pKa = 10.8DD187 pKa = 3.27PYY189 pKa = 9.69FTNDD193 pKa = 3.67LFDD196 pKa = 4.22SAWSKK201 pKa = 11.22GFVIIGKK208 pKa = 7.96VNWNTISYY216 pKa = 8.94VAGYY220 pKa = 8.07CQKK223 pKa = 11.14KK224 pKa = 10.28LFGEE228 pKa = 4.3VGKK231 pKa = 10.62KK232 pKa = 9.15IYY234 pKa = 9.81EE235 pKa = 4.2MKK237 pKa = 10.39GQEE240 pKa = 4.2PIYY243 pKa = 10.27ATMSRR248 pKa = 11.84NPGIGRR254 pKa = 11.84NYY256 pKa = 9.86YY257 pKa = 9.88EE258 pKa = 4.36EE259 pKa = 4.21NKK261 pKa = 8.35EE262 pKa = 3.96TIYY265 pKa = 10.69RR266 pKa = 11.84YY267 pKa = 10.74DD268 pKa = 3.38EE269 pKa = 4.56VITSKK274 pKa = 10.76GKK276 pKa = 9.06SVKK279 pKa = 9.79PPSYY283 pKa = 10.24FDD285 pKa = 3.53RR286 pKa = 11.84LMDD289 pKa = 3.91IGEE292 pKa = 4.31EE293 pKa = 3.81DD294 pKa = 4.12SIILKK299 pKa = 8.94EE300 pKa = 4.09LKK302 pKa = 9.73EE303 pKa = 3.92KK304 pKa = 10.69RR305 pKa = 11.84RR306 pKa = 11.84EE307 pKa = 3.92KK308 pKa = 11.06SNNALKK314 pKa = 10.36QKK316 pKa = 10.09YY317 pKa = 9.17AQTTLTIKK325 pKa = 10.3EE326 pKa = 3.86QLALEE331 pKa = 4.24EE332 pKa = 4.05RR333 pKa = 11.84TAVEE337 pKa = 4.21KK338 pKa = 10.73LKK340 pKa = 10.68KK341 pKa = 9.98YY342 pKa = 10.26NRR344 pKa = 11.84DD345 pKa = 3.55RR346 pKa = 11.84IGQQ349 pKa = 3.55
Molecular weight: 40.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1520 |
86 |
549 |
253.3 |
28.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.632 ± 1.727 | 0.855 ± 0.283 |
4.145 ± 0.646 | 8.947 ± 1.39 |
3.421 ± 0.954 | 6.645 ± 0.796 |
1.645 ± 0.384 | 6.579 ± 1.116 |
7.895 ± 2.042 | 5.329 ± 0.368 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.895 ± 0.355 | 5.658 ± 0.331 |
3.684 ± 0.795 | 4.276 ± 0.408 |
4.013 ± 0.324 | 8.289 ± 3.476 |
6.513 ± 0.993 | 3.355 ± 0.604 |
1.974 ± 0.344 | 6.25 ± 0.278 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
